https://github.com/althonos/proteinogenic
Chemical structure generation for protein sequences as SMILES string.
Science Score: 10.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
○CITATION.cff file
-
○codemeta.json file
-
○.zenodo.json file
-
○DOI references
-
○Academic publication links
-
✓Committers with academic emails
1 of 2 committers (50.0%) from academic institutions -
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (8.5%) to scientific vocabulary
Keywords
Repository
Chemical structure generation for protein sequences as SMILES string.
Basic Info
Statistics
- Stars: 4
- Watchers: 2
- Forks: 1
- Open Issues: 0
- Releases: 0
Topics
Metadata Files
README.md
proteinogenic 
Chemical structure generation for protein sequences as SMILES string.
🔌 Usage
This crate builds on top of purr, a crate providing
primitives for reading and writing SMILES.
Use the AminoAcid enum to encode the sequence residues, and build a SMILES
string with proteinogenic::smiles. For example with divergicin 750:
```rust extern crate proteinogenic;
let residues = "KGILGKLGVVQAGVDFVSGVWAGIKQSAKDHPNA" .chars() .map(proteinogenic::AminoAcid::from_char) .map(Result::unwrap); let s = proteinogenic::smiles(residues) .expect("failed to generate SMILES string"); ```
Additional modifications can be carried out by using a Peptide struct to
configure the rendering of the peptide. So far, disulfide bonds as well as
lanthionine bridges are supported, as well as head-to-tail cyclization.
For instance. we can generate the SMILES string of a
cyclotide such as
kalata B1:
```rust extern crate proteinogenic;
let residues = "GLPVCGETCVGGTCNTPGCTCSWPVCTRN" .chars() .map(proteinogenic::AminoAcid::from_char) .map(Result::unwrap);
let mut p = proteinogenic::Protein::new(residues); p.cyclization(proteinogenic::Cyclization::HeadToTail); p.crosslink(proteinogenic::CrossLink::Cystine(5, 19)).unwrap(); p.crosslink(proteinogenic::CrossLink::Cystine(9, 21)).unwrap(); p.cross_link(proteinogenic::CrossLink::Cystine(14, 26)).unwrap();
let s = p.smiles() .expect("failed to generate SMILES string"); ```
This SMILES string can be used in conjunction with other cheminformatics toolkits, for instance OpenBabel which can generate a PNG figure:
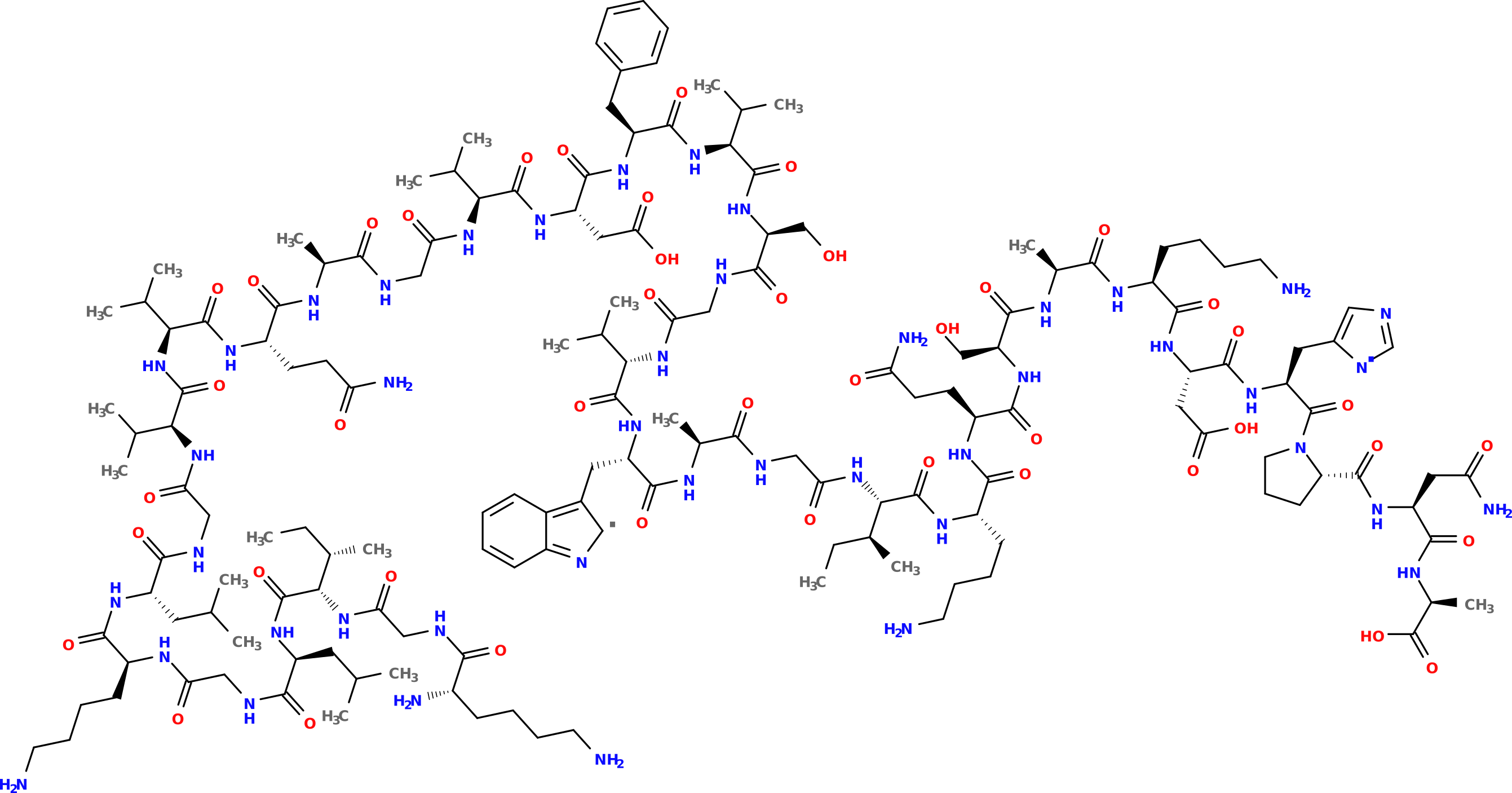
Note that proteinogenic is not limited to building a SMILES string; it can
actually use any purr::walk::Follower
implementor to generate an in-memory representation of a protein formula. If
your code is already compatible with purr, then you'll be able to use
protein sequences quite easily.
```rust extern crate proteinogenic; extern crate purr;
let sequence = "KGILGKLGVVQAGVDFVSGVWAGIKQSAKDHPNA"; let residues = sequence.chars() .map(proteinogenic::AminoAcid::from_char) .map(Result::unwrap);
let mut builder = purr::graph::Builder::new(); proteinogenic::visit(residues, &mut builder);
builder.build() .expect("failed to create a graph representation"); ```
The API is not yet stable, and may change to follow changes introduced by
purr or to improve the interface ergonomics.
💭 Feedback
⚠️ Issue Tracker
Found a bug ? Have an enhancement request ? Head over to the GitHub issue tracker if you need to report or ask something. If you are filing in on a bug, please include as much information as you can about the issue, and try to recreate the same bug in a simple, easily reproducible situation.
📋 Changelog
This project adheres to Semantic Versioning and provides a changelog in the Keep a Changelog format.
🔍 See Also
If you're a bioinformatician and a Rustacean, you may be interested in these other libraries:
uniprot.rs: Rust data structures for the UniProtKB databases.obofoundry.rs: Rust data structures for the OBO Foundry.fastobo: Rust parser and abstract syntax tree for Open Biomedical Ontologies.pubchem.rs: Rust data structures and API client for the PubChem API.
📜 License
This library is provided under the open-source MIT license.
This project was developed by Martin Larralde during his PhD project at the European Molecular Biology Laboratory in the Zeller team.
Owner
- Name: Martin Larralde
- Login: althonos
- Kind: user
- Location: Heidelberg, Germany
- Company: EMBL / LUMC, @zellerlab
- Twitter: althonos
- Repositories: 91
- Profile: https://github.com/althonos
PhD candidate in Bioinformatics, passionate about programming, SIMD-enthusiast, Pythonista, Rustacean. I write poems, and sometimes they are executable.
GitHub Events
Total
Last Year
Committers
Last synced: 10 months ago
Top Committers
| Name | Commits | |
|---|---|---|
| Martin Larralde | m****e@e****e | 34 |
| imgbot[bot] | 3****] | 1 |
Committer Domains (Top 20 + Academic)
Issues and Pull Requests
Last synced: 6 months ago
All Time
- Total issues: 0
- Total pull requests: 1
- Average time to close issues: N/A
- Average time to close pull requests: 22 minutes
- Total issue authors: 0
- Total pull request authors: 1
- Average comments per issue: 0
- Average comments per pull request: 1.0
- Merged pull requests: 1
- Bot issues: 0
- Bot pull requests: 1
Past Year
- Issues: 0
- Pull requests: 0
- Average time to close issues: N/A
- Average time to close pull requests: N/A
- Issue authors: 0
- Pull request authors: 0
- Average comments per issue: 0
- Average comments per pull request: 0
- Merged pull requests: 0
- Bot issues: 0
- Bot pull requests: 0
Top Authors
Issue Authors
Pull Request Authors
- imgbot[bot] (1)
Top Labels
Issue Labels
Pull Request Labels
Packages
- Total packages: 1
-
Total downloads:
- cargo 2,590 total
- Total dependent packages: 0
- Total dependent repositories: 0
- Total versions: 2
- Total maintainers: 1
crates.io: proteinogenic
Chemical structure generation for protein sequences as SMILES string
- Homepage: https://github.com/althonos/proteinogenic
- Documentation: https://docs.rs/proteinogenic/
- License: MIT
-
Latest release: 0.2.0
published about 4 years ago
Rankings
Maintainers (1)
Dependencies
- lazy_static 1.4.0 development
- pubchem 0.1.1 development
- purr 0.9.0







