Science Score: 77.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
✓CITATION.cff file
Found CITATION.cff file -
✓codemeta.json file
Found codemeta.json file -
✓.zenodo.json file
Found .zenodo.json file -
✓DOI references
Found 1 DOI reference(s) in README -
✓Academic publication links
Links to: arxiv.org -
✓Committers with academic emails
2 of 11 committers (18.2%) from academic institutions -
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (14.1%) to scientific vocabulary
Keywords
Keywords from Contributors
Repository
Spectroscopy lock application using RedPitaya
Basic Info
- Host: GitHub
- Owner: linien-org
- License: gpl-3.0
- Language: Python
- Default Branch: master
- Size: 5.27 MB
Statistics
- Stars: 92
- Watchers: 5
- Forks: 21
- Open Issues: 42
- Releases: 49
Topics
Metadata Files
README.md
Linien ‒ User-friendly locking of lasers using RedPitaya (STEMlab 125-14) that just works
![]()
User-friendly locking of lasers using RedPitaya (STEMlab 125-14) that just works. Linien aims to follow the UNIX philosophy of doing a single thing (locking using intelligent algorithms) very well. It was mainly developed for locking spectroscopy signals but may also be used for Pound-Drever-Hall or other lock-in techniques as well as simple PID operation. Linien is built with Python and Migen and is based on red pid.
Features
- All included: Sinusoidal modulation (up to 50 MHz), demodulation (1f to 5f), filtering and servo implemented on the FPGA.
- Client-server architecture: Autonomous operation on RedPitaya. One or multiple GUI clients or python clients can connect to the server.
- Autolock: Click and drag over a line, and Linien will automatically lock to it. This algorithm is built to be noise and jitter tolerant.
- IQ demodulation: Optimize demodulation phase in an instant
- Noise analysis: Record power spectral density (PSD) of the error signal for analyzing noise of the locked laser and for optimizing PID parameters
- Lock detection: Linien is capable of detecting loss of lock (temporarily disabled, use v0.3.2 if you rely in this feature
- Automatic relocking: if lock is lost, it relocks autonomously (temporarily disabled, use v0.3.2 if you rely in this feature)
- Machine learning is used to tune the spectroscopy parameters in order to optimize the signal
- Remote-controllable: the client libraries can be used to control or monitor the spectroscopy lock with Python.
- Combined FMS+MTS: Linien supports dual-channel spectroscopy that can be used to implement combined FMS+MTS
- Logging: Lock status and parameters can be logged to InfluxDB v2.
- Second integrator for slow control of piezo in an ECDL
- Additional analog outputs may be used using the GUI or python client (ANALOG_OUT 1, 2 and 3)
- 16 GPIO outputs may be programmed (e.g. for controlling other devices)
Getting started: Install Linien
Linien runs on Windows and Linux. For Windows users the standalone binaries containing the graphical user interface are recommended. These binaries run on your lab PC and contain everything to get Linien running on your RedPitaya.
Starting with Linien 2.0, only RedPitaya OS 2.x is supported. Linien 1.x works on RedPitaya OS but is no longer actively maintain.
Standalone binary
You can download standalone binaries for Windows on
the releases page (download the binary in the assets
section of the latest version). For Linux users, we recommend installation of linien-gui via pip.
Installation with pip
Linien is written for python 3 and can be installed using python\'s package manager pip:
bash
pip install linien-gui
The GUI can be started by calling
bash
linien
in a terminal (on both Linux and Windows).
In case you're only interested in the Python client and don't want to install the graphical application, you may use the linien-client package:
bash
pip install linien-client
Installation of the server on the RedPitaya
The easiest way to install the server component of Linien on the RedPitaya, is to use the graphical user interface. The first time you are connecting to the RedPitaya, the server is automatically installed.
In case you are using the linien-client, the server can be installed with
```python from linienclient.device import Device from linienclient.deploy import installremoteserver
device = Device(
host="rp-xxxxxx.local",
username="root",
password="root"
)
installremoteserver(device)
```
Finally, you can install the server manually, by connecting to the RedPitaya via SSH and then running
bash
pip install linien-server
The server can then be started as a systemd service by running
bash
linien-server start
on the RedPitaya. To check the status of the server, run
bash
linien-server status
For more options, run
bash
linien-server --help
Physical setup
The default setup looks like this:
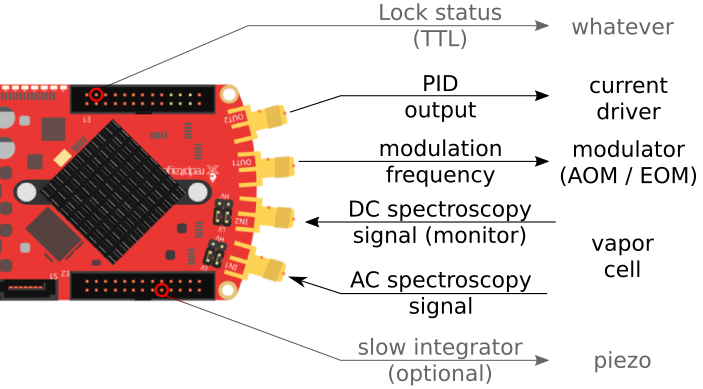
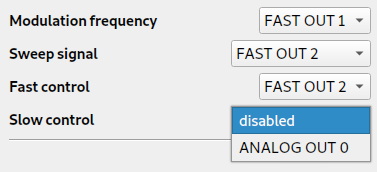
You can also configure Linien for different setups, e.g. if you want to have the modulation frequency and the control on the same output. Additionally, it is possible to set up a slow integrator on ANALOG OUT 0 (0 V to 1.8 V).
Using the application
First run: connecting to the RedPitaya
After launching Linien you should supply details of your RedPitaya. Its host address is usually given by
rp-XXXXXX.local, where XXXXXX are the last 6 digits of the device's MAC address. You will find them on a sticker on the ethernet port:

| :exclamation: If connecting using host name fails, try using RP's IP address instead | |--------------------------------------------------------------------------------------|
Default value for user name and password is root (but you should probably change the password...).
When connecting to a RedPitaya for the first time, the Linien offers you to install the server component. Please note that this requires internet access on the RedPitaya (LAN access is not sufficient).
Once server libraries are installed, Linien will automatically run the server and connect to it. There's no need ever to start or stop anything on the server manually as the client takes care of this.
The server now operates autonomously: closing the client application doesn't have any influence on the lock status. You may also start multiple clients connecting to the same server.
Setting things up
The first thing to set up is the configuration of input and output signals:
Connect your AC spectroscopy signal to FAST IN 1. If you also want to monitor the DC spectroscopy signal, connect it to FAST IN 2.
Then, adapt the output signals to your needs:

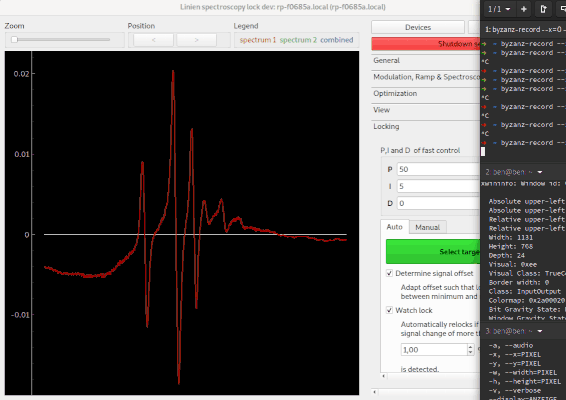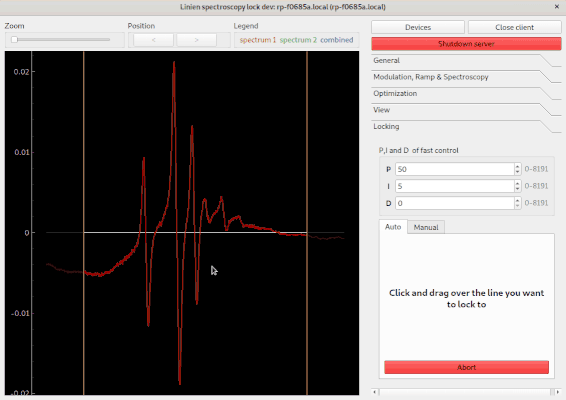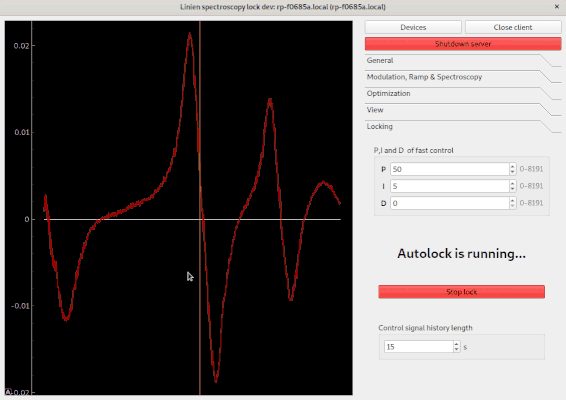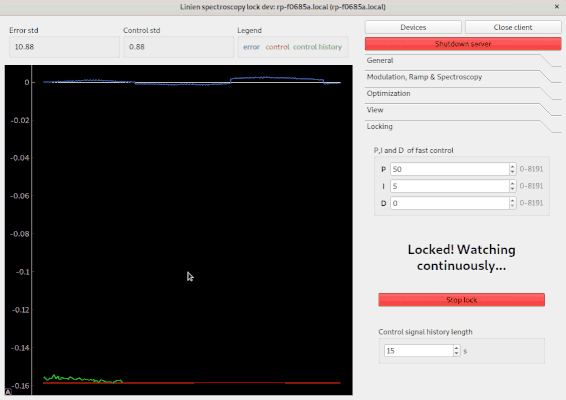
When you're done, head over to Modulation, Sweep & Spectroscopy to configure modulation frequency and amplitude. Once your setup is working, you should see something like this:
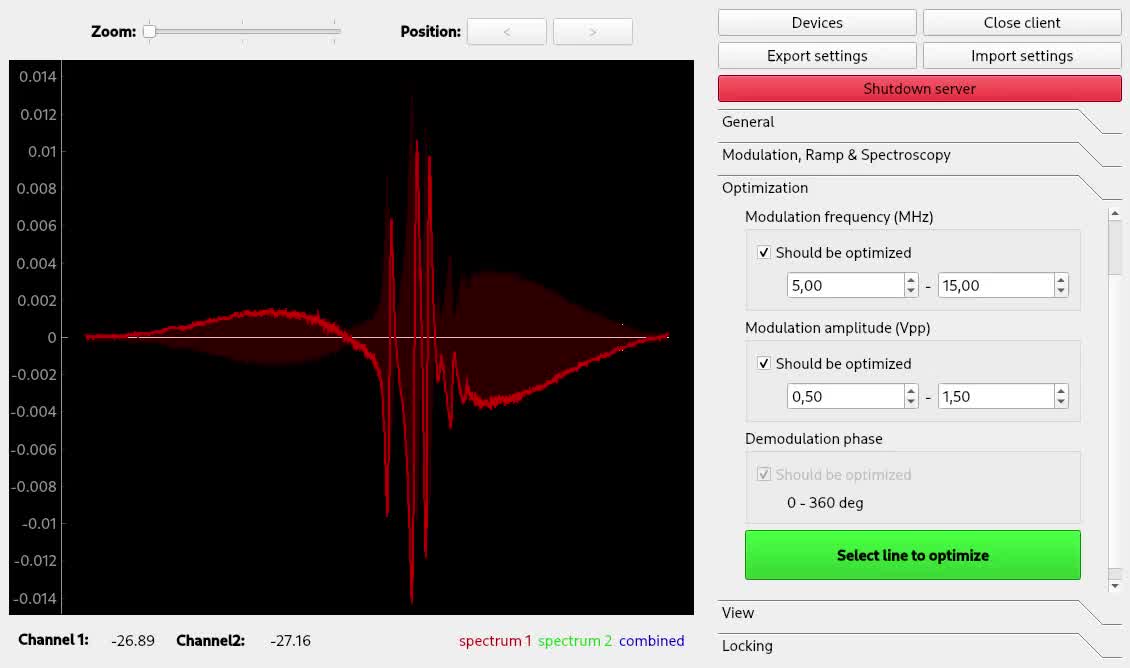
The bright red line is the demodulated spectroscopy signal. The dark red area is the signal strength obtained by iq demodulation, i.e. the demodulation signal obtained when demodulating in phase at this point.
PID-only mode
PID-only mode is intended for bare PID operation (no demodulation or filtering), bypassing most of the FPGA functionality. If enabled, the signal flow is FAST IN 1 → PID → FAST OUT 2. This is useful, if aiming for a high control bandwidth: PID-only mode reduces propagation delay from 320 ns to 125 ns which may make a difference when phase-locking lasers.
Optimization of spectroscopy parameters using machine learning
Linien may use machine learning to maximize the slope of a line. As for the autolock, click and drag over the line you want to optimize. Then, the line is centered and the optimization starts. Please note that this only works if initially a distinguished zero-crossing is visible.
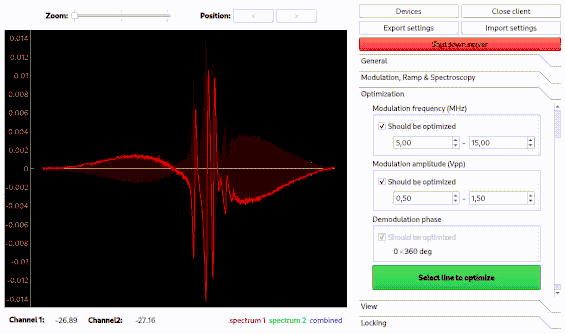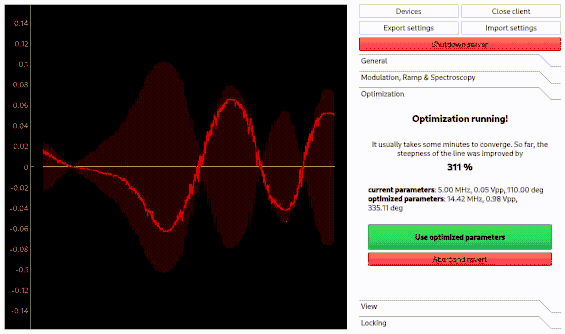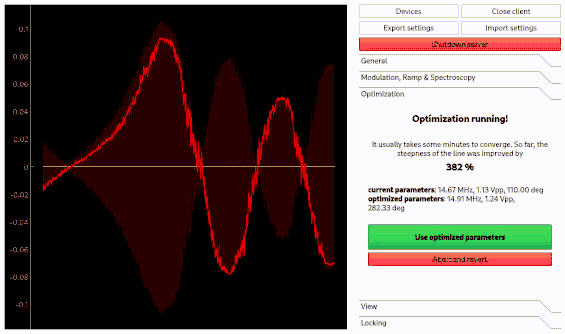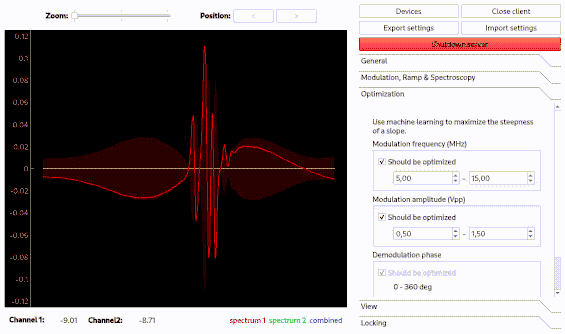
Using the autolock
In order to use the autolock, enter some PID parameters first. Note that the sign of the parameters is determined automatically. After clicking the green button, you can select the line you want to lock to by clicking and dragging over it: your selection should contain both extrema of the line. The autolock will then center this line, decrease the scan range and try to lock to the middle between minimum and maximum contained in your selection.

The following options are available: * Determine signal offset: If this checkbox is ticked, the autolock will adapt the y-offset of the signal such that the middle between minimum and maximum is at the zero crossing. This is especially useful if you have a large background signal (e.g. the Doppler background in FMS spectroscopy). * Check lock: Directly after turning on the lock, the control signal is investigated. If it shifts too much, the lock is assumed to have failed. * Watch lock: This option tells the Linien to continuously watch the control signal when the laser is locked. If steep changes are detected, a relock is initiated.
If you experience trouble with the autolock, this is most likely due to a bad signal-to-noise ratio or strong laser jitter.
Autolock algorithms
Linien implements two different autolock algorithms:
- Robust mode: this algorithm runs on FPGA and analyzes the peak shapes in order to turn on the lock at the right sweep position. It is able to cope with a high amount of jitter as it runs completely on the FPGA, i.e. no delays due to communication between CPU and FPGA occur.
- Simple mode: this algorithm uses a simple calculation of auto-correlation on the CPU which is then used to specify at which point of the sweep the lock should start. This algorithm is less complex than the first one and may be used if you experience problems with jitter-tolerant mode. As it requires some communication between CPU and FPGA which causes some delay, it may have problems if the line jitters a lot.
By default, auto-detect mode is used: this mode chooses an algorithm based on the amount of jitter.
Using the manual lock
If you have problems with the autolock, you may also lock manually. Activate the Manual tab and use the controls in the top (Zoom and Position) to center the line you want to lock to. Choose whether the target slope is rising or falling and click the green button.
Logging
Linien has to option to log the lock status and parameters to an InfluxDB. Currently, only InfluxDB 2.x is supported. Logging can be configured via the Logging menu in the Linien GUI, but logging will continue even if the client is closed. Time stamps of the data points are determined by the InfluxDB, not by the RedPitaya. If updating/checking the InfluxDB credentials fails, there is additional information in the tool-tip of the fail indicator ❌.
The parameter names are documented in parameters.py. The signal_stats parameter does contain statistics of the input and output signals, e.g. control_signal_mean or monitor_signal_max.
Transfer function
Transfer function of the PID is given by
L(f) = kp + ki / f + kd * f
with kp=P/4096, ki=I/0.1s and kd=D / (2**6 * 125e6).
Note that this equation does not account for filtering before the PID (cf. Modulation, Sweep & Spectroscopy tab).
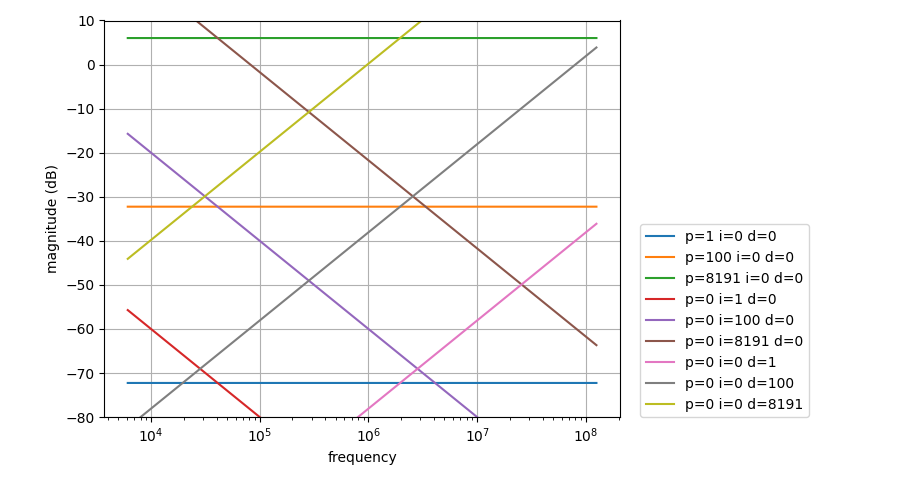
Scripting interface
In addition to the GUI, Linien can also be controlled using Python. For that purpose, installation via pip is required (see above).
Then, you should start the Linien server on your RedPitaya. This can be done by running the GUI client and connecting to the device (see above). Alternatively, the connect method of LinienClient has the option autostart_server.
Once the server is up and running, you can connect using python: ```python from linienclient.device import Device from linienclient.connection import LinienClient from liniencommon.common import MHz, Vpp, ANALOGOUT_V
dev = Device(
host="rp-xxxxxx.local",
username="root",
password="root"
)
c = LinienClient(dev)
c.connect(autostartserver=True, useparameter_cache=True)
read out the modulation frequency
print(c.parameters.modulation_frequency.value / MHz)
have a look at https://github.com/linien-org/linien/blob/master/linien/server/parameters.py
for a documentation of all parameters that can be accessed and modified
set modulation amplitude
c.parameters.modulation_amplitude.value = 1 * Vpp
in the line above, we set a parameter. This is not written directly to the
FPGA, though. In order to do this, we have to call write_registers():
c.connection.root.write_registers()
additionally set ANALOGOUT1 to 1.2 volts DC (you can use this to control other devices of your experiment)
c.parameters.analogout1.value = 1.2 / ANALOGOUTV
GPIO outputs can also be set
each bit corresponds to a pin
Example: enable all N pins and disable all P pins
c.parameters.gpionout.value = 0b11111111 c.parameters.gpiopout.value = 0b00000000
Example: enable the N pins 1-4 and disable N pins 5-8
c.parameters.gpionout.value = 0b11110000 # 4 on, 4 off
Example: enable every second P pin
c.parameters.gpiopout.value = 0b01010101 # 4 on, 4 off
again, we have to call write_registers in order to write the data to the FPGA
c.connection.root.write_registers()
it is also possible to set up a callback function that is called whenever a
parameter changes (remember to call check_for_changed_parameters() periodically)
def callback(value):
# this function is called whenever my_param changes on the server.
# note that this only works if check_for_changed_parameters is called from
# time to time as this function is responsible for checking for
# changed parameters.
print('parameter arrived!', value)
c.parameters.modulationamplitude.addcallback(callback)
from time import sleep for i in range(10): c.parameters.checkforchangedparameters() if i == 2: c.parameters.modulationamplitude.value = 0.1 * Vpp sleep(.1)
plot control and error signal
import pickle from matplotlib import pyplot as plt plotdata = pickle.loads(c.parameters.toplot.value)
depending on the status (locked / unlocked), different signals are available
print(plot_data.keys())
if unlocked, signal1 and signal2 contain the error signal of channel 1 and 2
if the laser is locked, they contain error signal and control signal.
if c.parameters.lock.value: plt.title('laser is locked!') plt.plot(plotdata['controlsignal'], label='control signal') plt.plot(plotdata['errorsignal'], label='error signal') else: plt.title('laser is sweeping!') plt.plot(plotdata['errorsignal1'], label='error signal channel 1') plt.plot(plotdata['errorsignal2'], label='error signal channel 2')
plt.legend() plt.show() ```
For a full list of parameters that can be controlled or accessed have a
look at
parameters.py. Remember that changed parameters are not written to the FPGA unless c.connection.root.write_registers() is called.
Run the autolock
The script below shows an example of how to run the autolock using the scripting interface:
```python import pickle import numpy as np
from linienclient.connection import LinienClient from liniencommon.common import FAST_AUTOLOCK
from matplotlib import pyplot as plt from time import sleep
c = LinienClient( host="rp-xxxxxx.local", username="root", password="root" ) c.connect(autostartserver=True, useparameter_cache=True)
c.parameters.autolockmodepreference.value = FAST_AUTOLOCK
def waitforlockstatus(shouldbelocked): """A helper function that waits until the laser is locked or unlocked.""" counter = 0 while True: toplot = pickle.loads(c.parameters.toplot.value) islocked = "errorsignal" in toplot
if is_locked == should_be_locked:
break
counter += 1
if counter > 10:
raise Exception("waited too long")
sleep(1)
turn of the lock (if it is running)
c.connection.root.start_sweep()
wait until the laser is unlocked (if required)
waitforlock_status(False)
we record a reference spectrum
toplot = pickle.loads(c.parameters.toplot.value) errorsignal = toplot["errorsignal1"]
we plot the reference spectrum and ask the user where the target line is
plt.plot(errorsignal) plt.plot(toplot["monitor_signal"]) plt.show()
print("Please specify the position of the target line. ") x0 = int(input("enter index of a point that is on the left side of the target line: ")) x1 = int(input("enter index of a point that is on the right side of the target line: "))
show the lock point again
plt.axvline(x0, color="r") plt.axvline(x1, color="r") plt.plot(error_signal) plt.show()
turn on the lock
c.connection.root.startautolock(x0, x1, pickle.dumps(errorsignal))
wait until the laser is actually locked
try: waitforlock_status(True) print("locking the laser worked \o/") except Exception: print("locking the laser failed :(")
```
Updating Linien
Before installing a new version of Linien, open the previously installed client and click the "Shutdown server" button. Don't worry, your settings and parameters will be saved. Then you may install the latest client on your local PC as described in the getting started section above. The next time you connect to RedPitaya, Linien will install the matching server version.
Development
Information about development can be found in the wiki.
FAQs
How to update to a new version?
There's no need to install anything on RedPitaya manually. Run the new version of Linien on your computer and connect to RedPitaya. You will see a dialog that allows you to install the corresponding server component.
Can I run Linien and the RedPitaya web application / scpi interface at the same time?
No, this is not possible as Linien relies on a customized FPGA bitstream.
What control bandwidth is achievable with Linien?
The propagation delay is roughly 320 ns in normal mode and 125 ns in PID-only mode.
Why do ethernet LEDs of RedPitaya stop blinking when Linien is running?
Ethernet LED blinking was found to impact analog outputs of RedPitaya. As this may impact lock stability, Linien disables ethernet LED blinking when starting.
If you want to re-enable the LEDs, just stop the Linien server or restart your RedPitaya.
Troubleshooting
Connection problems
If the client fails to connect to a RedPitaya, first check whether you can ping it by executing
bash
ping rp-f0xxxx.local
in a command line. If this works, check whether you can connect via SSH:
bash
ssh rp-f0xxxx.local
on the command line. If this is successful, in order to check whether the
linien-server is running, check that the systemd service is running. This can be done
by executing linien-server status. Errors will also be displayed. Please provide the
output if you are reporting an issue
related to connection problems. Debugging info will also be stored in /root/.local/share/linien/linien.log.
Possible conflict with openSSH
Note that there might be issues if openSSH server is running on the client, see here.
Updating or installing fails
- make sure that your RedPitaya is connected to the internet
- if the orange LED stops blinking and RedPitaya becomes unresponsive, your SD card is probably faulty
Citation
If you are using Linien for your scientific work, please cite us as follows:
@article{Wiegand2022,
author = {B. Wiegand and B. Leykauf and R. Jördens and M. Krutzik},
doi = {10.1063/5.0090384},
issn = {10897623},
issue = {6},
journal = {Review of Scientific Instruments},
month = {6},
pmid = {35778046},
title = {Linien: A versatile, user-friendly, open-source FPGA-based tool for frequency stabilization and spectroscopy parameter optimization},
volume = {93},
year = {2022},
}
License
Linien ‒ User-friendly locking of lasers using RedPitaya (STEMlab 125-14) that just works.
Copyright © 2014-2015 Robert Jördens\ Copyright © 2018-2022 Benjamin Wiegand\ Copyright © 2021-2024 Bastian Leykauf\ Copyright © 2022 Christian Freier\ Copyright © 2023-2024 Doron Behar\
Linien is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation, either version 3 of the License, or (at your option) any later version.
Linien is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.
You should have received a copy of the GNU General Public License along with Linien. If not, see https://www.gnu.org/licenses/.
Acknowledgements
Development mainly takes place at Humboldt University of Berlin.
This work is supported by the German Space Agency (DLR) with funds provided by the Federal Ministry of Economics and Technology (BMWi) under grant number DLR50WM2066.
See Also
- RedPID: the basis of Linien
Owner
- Name: Linien
- Login: linien-org
- Kind: organization
- Location: Germany
- Repositories: 3
- Profile: https://github.com/linien-org
User-friendly locking of lasers using RedPitaya (STEMlab 125-14) that just works.
Citation (CITATION.cff)
cff-version: 1.2.0
message: If you are using Linien as part of a scientific publication, please cite our article.
authors:
- given-names: Benjamin
family-names: Wiegand
email: bwiegand@physik.hu-berlin.de
affiliation: Institut für Physik, Humboldt-Universität zu Berlin
orcid: "https://orcid.org/0000-0001-6549-2307"
- family-names: Leykauf
given-names: Bastian
email: leykauf@physik.hu-berlin.de
affiliation: Institut für Physik, Humboldt-Universität zu Berlin
orcid: "https://orcid.org/0000-0002-4630-0602"
- family-names: Jördens
given-names: Robert
affiliation: '"QUARTIQ GmbH'
orcid: "https://orcid.org/0000-0001-5333-0562"
preferred-citation:
type: article
title: >-
Linien: A versatile, user-friendly, open-source FPGA-based tool for frequency
stabilization and spectroscopy parameter optimization
authors:
- given-names: Benjamin
family-names: Wiegand
email: bwiegand@physik.hu-berlin.de
affiliation: >-
Institut für Physik, Humboldt-Universität zu
Berlin
orcid: "https://orcid.org/0000-0001-6549-2307"
- family-names: Leykauf
given-names: Bastian
email: leykauf@physik.hu-berlin.de
affiliation: >-
Institut für Physik, Humboldt-Universität zu
Berlin
orcid: "https://orcid.org/0000-0002-4630-0602"
- family-names: Jördens
given-names: Robert
affiliation: '"QUARTIQ GmbH'
orcid: "https://orcid.org/0000-0001-5333-0562"
- family-names: Krutzik
given-names: Markus
affiliation: >-
Institut für Physik, Humboldt-Universität zu
Berlin & Ferdinand-Braun-Institut gGmbH,
Leibniz-Institut für Höchstfrequenztechnik
doi: "10.1063/5.0090384"
url: "https://doi.org/10.1063/5.0090384"
journal: "Review of Scientific Instruments"
volume: 93
issue: 6
start: "063001"
year: 2022
GitHub Events
Total
- Issues event: 6
- Watch event: 24
- Issue comment event: 3
- Push event: 18
- Pull request event: 1
- Fork event: 2
Last Year
- Issues event: 6
- Watch event: 24
- Issue comment event: 3
- Push event: 18
- Pull request event: 1
- Fork event: 2
Committers
Last synced: over 1 year ago
Top Committers
| Name | Commits | |
|---|---|---|
| Bastian Leykauf | l****f@p****e | 1,079 |
| Robert Jordens | j****s@g****m | 287 |
| Benjamin Wiegand | h****e@z****m | 258 |
| Benjamin Wiegand | h****e@p****e | 228 |
| pre-commit-ci[bot] | 6****] | 31 |
| oanton | o****n@p****e | 5 |
| dependabot[bot] | 4****] | 5 |
| Doron Behar | d****r@g****m | 4 |
| root | r****t@r****p | 3 |
| Christian Freier | c****r@g****m | 2 |
| Florent Kermarrec | f****t@e****r | 1 |
Committer Domains (Top 20 + Academic)
Issues and Pull Requests
Last synced: 6 months ago
All Time
- Total issues: 84
- Total pull requests: 109
- Average time to close issues: 8 months
- Average time to close pull requests: about 1 month
- Total issue authors: 36
- Total pull request authors: 7
- Average comments per issue: 2.21
- Average comments per pull request: 0.57
- Merged pull requests: 91
- Bot issues: 0
- Bot pull requests: 22
Past Year
- Issues: 8
- Pull requests: 2
- Average time to close issues: 17 minutes
- Average time to close pull requests: 4 days
- Issue authors: 8
- Pull request authors: 1
- Average comments per issue: 0.5
- Average comments per pull request: 0.0
- Merged pull requests: 1
- Bot issues: 0
- Bot pull requests: 2
Top Authors
Issue Authors
- bleykauf (27)
- hermitdemschoenenleben (7)
- doronbehar (4)
- martinssonh (3)
- NekSadam (3)
- wwcphy (3)
- systemofapwne (3)
- Patschke (2)
- jmatyas (2)
- ZhimingShi (2)
- ograsdijk (2)
- SoraUmika (2)
- fartmano (2)
- andrewvh4 (2)
- gregmoille (1)
Pull Request Authors
- bleykauf (91)
- pre-commit-ci[bot] (22)
- doronbehar (10)
- dependabot[bot] (5)
- hermitdemschoenenleben (4)
- kongmunist (2)
- cmf84 (1)
Top Labels
Issue Labels
Pull Request Labels
Packages
- Total packages: 6
-
Total downloads:
- pypi 1,093 last-month
-
Total dependent packages: 6
(may contain duplicates) -
Total dependent repositories: 3
(may contain duplicates) - Total versions: 276
- Total maintainers: 2
pypi.org: linien-server
Server components of the Linien spectroscopy lock application.
- Homepage: https://github.com/linien-org/linien
- Documentation: https://linien-server.readthedocs.io/
- License: GNU General Public License v3 (GPLv3)
-
Latest release: 2.1.0
published over 1 year ago
Rankings
Maintainers (2)
pypi.org: linien
Spectroscopy lock application using RedPitaya
- Homepage: https://github.com/linien-org/linien
- Documentation: https://linien.readthedocs.io/
- License: GNU General Public License v3 (GPLv3)
-
Latest release: 0.5.3
published almost 4 years ago
Rankings
Maintainers (2)
pypi.org: linien-python-client
Python client for linien spectroscopy lock
- Homepage: https://github.com/linien-org/linien/
- Documentation: https://linien-python-client.readthedocs.io/
- License: GNU General Public License v3 (GPLv3)
-
Latest release: 0.5.3
published almost 4 years ago
Rankings
Maintainers (2)
pypi.org: linien-common
Shared components of the Linien spectroscopy lock application.
- Homepage: https://github.com/linien-org/linien
- Documentation: https://linien-common.readthedocs.io/
- License: GNU General Public License v3 (GPLv3)
-
Latest release: 2.1.0
published over 1 year ago
Rankings
Maintainers (1)
pypi.org: linien-client
Client components of the Linien spectroscopy lock application.
- Homepage: https://github.com/linien-org/linien/
- Documentation: https://linien-client.readthedocs.io/
- License: GNU General Public License v3 (GPLv3)
-
Latest release: 2.1.0
published over 1 year ago
Rankings
Maintainers (1)
pypi.org: linien-gui
Graphical user interface of the Linien spectroscopy lock application.
- Homepage: https://github.com/linien-org/linien
- Documentation: https://linien-gui.readthedocs.io/
- License: GNU General Public License v3 (GPLv3)
-
Latest release: 2.1.0
published over 1 year ago
Rankings
Maintainers (1)
Dependencies
- numpy ==1.21.0
- paramiko ==2.10.1
- plumbum ==1.6.9
- rpyc ==4.1.5
- scipy ==1.4.1
- uuid ==1.30
- black ==22.3.0 development
- cma ==3.0.3 development
- flake8 * development
- matplotlib * development
- migen ==0.9.2 development
- pre-commit * development
- pyinstaller ==3.6 development
- pylpsd ==0.1.3 development
- pytest * development
- pytest-plt * development
- twine * development
- PyNaCl ==1.4.0
- PyQt5 ==5.15.0
- PyQt5-sip ==12.8.0
- altgraph ==0.17
- appdirs ==1.4.4
- bcrypt ==3.2.0
- cffi ==1.14.2
- click ==8.1.2
- cryptography ==3.3.2
- future ==0.18.2
- numpy ==1.21.0
- packaging ==21.3
- paramiko ==2.10.1
- pefile ==2019.4.18
- plumbum ==1.6.9
- pycparser ==2.20
- pyqtgraph ==0.11.0
- pywin32-ctypes ==0.2.0
- rpyc ==4.1.5
- scipy ==1.4.1
- six ==1.15.0
- superqt ==0.2.3
- uuid ==1.30
- click ==7.1.2
- cma ==3.0.3
- myhdl ==0.11
- numpy ==1.11.0
- plumbum ==1.6.9
- pylpsd ==0.1.4
- rpyc ==4.1.5
- scipy ==0.17.0
- JackMcKew/pyinstaller-action-linux main composite
- actions/checkout v2 composite
- actions/upload-artifact v2 composite
- actions/checkout v3 composite
- actions/setup-python v4 composite
- actions/upload-artifact v3 composite
- JackMcKew/pyinstaller-action-windows main composite
- actions/checkout v2 composite
- actions/upload-artifact v2 composite
- JackMcKew/pyinstaller-action-windows main composite
- actions/checkout v2 composite
- actions/setup-python v2 composite
- actions/upload-artifact v2 composite


