abspy
abspy: A Python package for 3D adaptive binary space partitioning and modeling - Published in JOSS (2025)
Science Score: 93.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
○CITATION.cff file
-
✓codemeta.json file
Found codemeta.json file -
✓.zenodo.json file
Found .zenodo.json file -
✓DOI references
Found 6 DOI reference(s) in README and JOSS metadata -
✓Academic publication links
Links to: sciencedirect.com -
○Committers with academic emails
-
○Institutional organization owner
-
✓JOSS paper metadata
Published in Journal of Open Source Software
Keywords
Scientific Fields
Repository
3D adaptive binary space partitioning and beyond
Basic Info
- Host: GitHub
- Owner: chenzhaiyu
- License: mit
- Language: Python
- Default Branch: main
- Homepage: https://abspy.readthedocs.io/
- Size: 13.6 MB
Statistics
- Stars: 89
- Watchers: 4
- Forks: 13
- Open Issues: 1
- Releases: 6
Topics
Metadata Files
README.md

Introduction
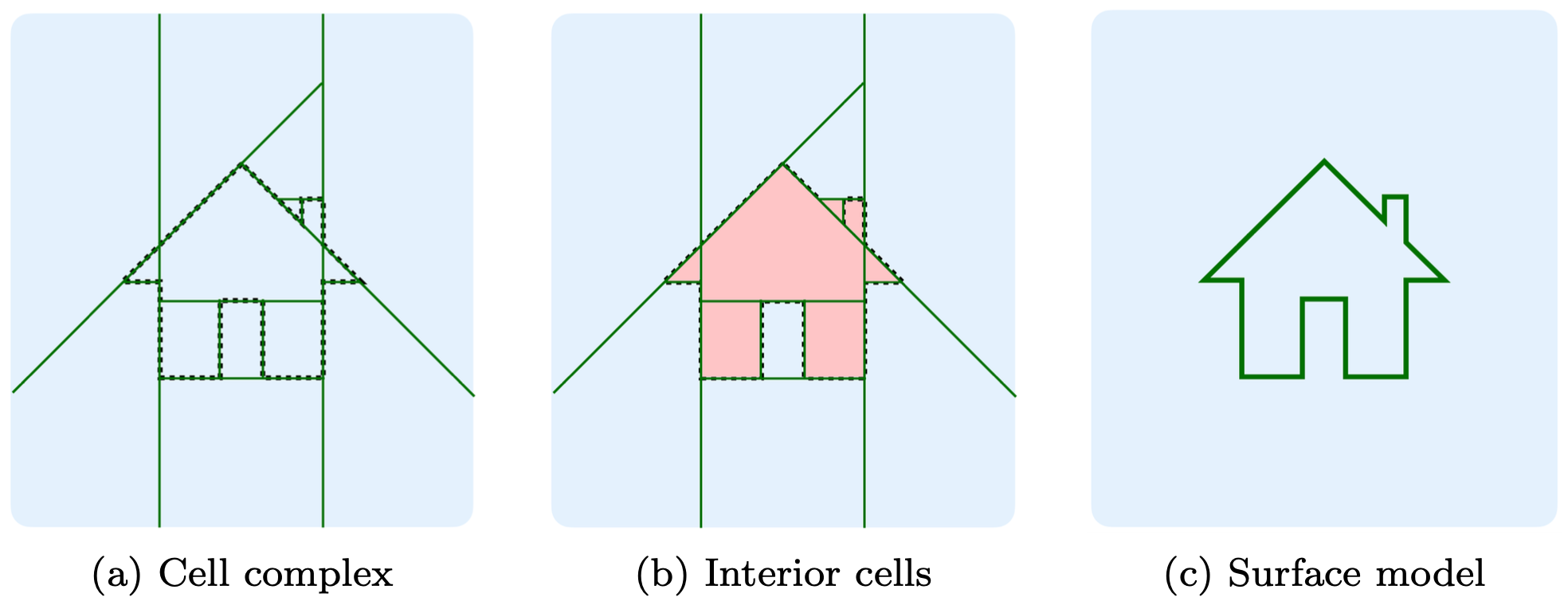
abspy is a Python tool for 3D adaptive binary space partitioning and beyond. It adaptively partitions ambient 3D space into a linear cell complex using planar primitives, dynamically generating an adjacency graph in the process. Designed primarily for compact surface reconstruction, abspy also supports a range of other applications.

Key features
- Manipulation of planar primitives from point cloud or reference mesh
- Linear cell complex creation with adaptive binary space partitioning (a-BSP)
- Dynamic BSP-tree (NetworkX graph) updated locally upon primitive insertion
- Support of polygonal surface reconstruction with graph cut
- Compatible data structure with Easy3D on point cloud, primitive, mesh and cell complex
- Robust spatial operations underpinned by the rational ring from SageMath's exact kernel
Installation
All-in-one installation
Create a conda environment with the latest abspy release and all its dependencies installed:
bash
git clone https://github.com/chenzhaiyu/abspy && cd abspy
conda env create -f environment.yml && conda activate abspy
Manual installation
Still easy! Create a conda environment and enter it:
bash
conda create --name abspy python=3.11 && conda activate abspy
Install the dependencies:
```bash
You may replace conda with mamba for faster package parsing
conda install mamba -c conda-forge
conda install -c conda-forge networkx numpy tqdm scikit-learn matplotlib colorlog scipy trimesh rtree pyglet sage=10.2 ```
Preferably, the latest abspy release can be found and installed via PyPI:
bash
pip install abspy
Otherwise, you can install the latest version locally:
bash
git clone https://github.com/chenzhaiyu/abspy && cd abspy
pip install .
Quick start
Example 1 - Reconstruction from point cloud
The example loads a point cloud to VertexGroup (.vg), partitions ambient space into a cell complex, creates the adjacency graph, and extracts the object's outer surface.
```python from abspy import VertexGroup, AdjacencyGraph, CellComplex
load a point cloud in VertexGroup
vertexgroup = VertexGroup(filepath='tutorials/data/testpoints.vg')
normalise point cloud
vertexgroup.normalisetocentroidand_scale()
initialise cell complex
cellcomplex = CellComplex(vertexgroup.planes, vertexgroup.aabbs, vertexgroup.obbs, vertexgroup.pointsgrouped, buildgraph=True, additionalplanes=None)
refine planar primitives
cellcomplex.refineplanes()
prioritise certain planes (e.g., vertical ones)
cellcomplex.prioritiseplanes(prioritise_verticals=False)
construct cell complex
cell_complex.construct()
print info about cell complex
cellcomplex.printinfo()
cells inside reference mesh
cellsinmesh = cellcomplex.cellsinmesh('tutorials/data/testmesh.ply')
build adjacency graph from cell complex
adjacencygraph = AdjacencyGraph(cellcomplex.graph)
calculate volumes
volumes = cellcomplex.volumes(multiplier=10e5) volumes = [0.1 if i in cellsin_mesh else vol for i, vol in enumerate(volumes)]
assign graph weights
adjacencygraph.assignweightstonlinks(cellcomplex.cells, attribute='areaoverlap', factor=0.000, cacheinterfaces=True) adjacencygraph.assignweightstostlinks(adjacencygraph.to_dict(volumes))
perform graph cut to extract surface
, _ = adjacencygraph.cut()
save surface model to an OBJ file
adjacencygraph.savesurface_obj('tutorials/output/surface.obj', engine='mesh') ```
Example 2 - Convex decomposition from mesh
The example loads a mesh to VertexGroupReference, partitions ambient space into a cell complex, identifies cells inside reference mesh, and visualizes the cells.
```python from abspy import VertexGroupReference vertexgroupreference = VertexGroupReference(filepath='tutorials/data/test_mesh.ply')
initialise cell complex
cellcomplex = CellComplex(vertexgroupreference.planes, vertexgroupreference.aabbs, vertexgroupreference.obbs, buildgraph=True)
construct cell complex
cell_complex.construct()
cells inside reference mesh
cellsinmesh = cellcomplex.cellsinmesh('tutorials/data/testmesh.ply', engine='distance')
save cell complex file
cell_complex.save('tutorials/output/complex.cc')
visualise the inside cells
if len(cellsinmesh): cellcomplex.visualise(indicescells=cellsinmesh) ```
Please find the usage of abspy at API reference, with self-explanatory examples in ./tutorials.
For the data structure of a .vg/.bvg file, please refer to VertexGroup.
Testing abspy
To run the test suite, first install pytest, and execute all tests:
bash
pytest
Contributing to abspy
Please see the Contribution Guide for more information.
FAQ
- How can I install abspy on Windows
For Windows users, you may need to build SageMath from source or install all other dependencies into a pre-built SageMath environment. Otherwise, virtualization with docker may come to the rescue.
- How can I use abspy for surface reconstruction?
As shown in Example 1, the surface is defined between adjacent cells where one is inside and the other outside. For more information, please refer to Points2Poly which integrates abspy with deep implicit fields, and PolyGNN which learns a piecewise planar occupancy function supported by abspy, for 3D building reconstruction.
License
See the MIT license for details.
Citation
If you use abspy in a scientific work, please consider citing the paper:
bibtex
@article{chen2022points2poly,
title = {Reconstructing compact building models from point clouds using deep implicit fields},
journal = {ISPRS Journal of Photogrammetry and Remote Sensing},
volume = {194},
pages = {58-73},
year = {2022},
issn = {0924-2716},
doi = {https://doi.org/10.1016/j.isprsjprs.2022.09.017},
url = {https://www.sciencedirect.com/science/article/pii/S0924271622002611},
author = {Zhaiyu Chen and Hugo Ledoux and Seyran Khademi and Liangliang Nan}
}
To cite the abspy software package itself, please use:
bibtex
@article{chen2025abspy,
title = {abspy: A Python package for 3D adaptive binary space partitioning and modeling},
journal = {Journal of Open Source Software},
volume = {10},
number = {110},
pages = {7946},
year = {2025},
doi = {10.21105/joss.07946},
url = {https://doi.org/10.21105/joss.07946},
publisher = {The Open Journal},
author = {Zhaiyu Chen},
}
Owner
- Name: Zhaiyu Chen
- Login: chenzhaiyu
- Kind: user
- Location: Munich, Germany
- Company: Technical University of Munich
- Website: chenzhaiyu.com
- Repositories: 32
- Profile: https://github.com/chenzhaiyu
JOSS Publication
abspy: A Python package for 3D adaptive binary space partitioning and modeling
Tags
3D modeling point cloud binary space partitioning surface reconstructionGitHub Events
Total
- Create event: 7
- Issues event: 21
- Release event: 2
- Watch event: 17
- Delete event: 2
- Issue comment event: 24
- Push event: 64
- Pull request event: 3
- Fork event: 1
Last Year
- Create event: 7
- Issues event: 21
- Release event: 2
- Watch event: 17
- Delete event: 2
- Issue comment event: 24
- Push event: 64
- Pull request event: 3
- Fork event: 1
Committers
Last synced: about 2 years ago
Top Committers
| Name | Commits | |
|---|---|---|
| Zhaiyu Chen | z****n@o****m | 54 |
| chenzhaiyu | 2****u | 39 |
| Yuqing Wang | y****g@g****m | 1 |
Committer Domains (Top 20 + Academic)
Issues and Pull Requests
Last synced: 6 months ago
All Time
- Total issues: 22
- Total pull requests: 14
- Average time to close issues: 5 months
- Average time to close pull requests: 1 day
- Total issue authors: 8
- Total pull request authors: 2
- Average comments per issue: 2.32
- Average comments per pull request: 0.29
- Merged pull requests: 12
- Bot issues: 0
- Bot pull requests: 0
Past Year
- Issues: 11
- Pull requests: 4
- Average time to close issues: 10 days
- Average time to close pull requests: 4 minutes
- Issue authors: 3
- Pull request authors: 1
- Average comments per issue: 1.45
- Average comments per pull request: 0.0
- Merged pull requests: 3
- Bot issues: 0
- Bot pull requests: 0
Top Authors
Issue Authors
- chenzhaiyu (7)
- tuelwer (6)
- preetishkakkar (4)
- fangxunyes (1)
- raphaelsulzer (1)
- yqf2000119 (1)
- matthewfeickert (1)
- zuixiaosanlang (1)
Pull Request Authors
- chenzhaiyu (13)
Top Labels
Issue Labels
Pull Request Labels
Packages
- Total packages: 1
-
Total downloads:
- pypi 364 last-month
- Total dependent packages: 0
- Total dependent repositories: 1
- Total versions: 21
- Total maintainers: 1
pypi.org: abspy
A Python tool for 3D adaptive binary space partitioning and beyond
- Homepage: https://github.com/chenzhaiyu/abspy
- Documentation: https://abspy.readthedocs.io/
- License: 'MIT'
-
Latest release: 0.3.0
published 9 months ago
Rankings
Maintainers (1)
Dependencies
- actions/checkout v3 composite
- actions/setup-python v3 composite
- pypa/gh-action-pypi-publish 27b31702a0e7fc50959f5ad993c78deac1bdfc29 composite
- abspy *
- attrs ==21.4.0
- myst_parser ==0.16.1
- sphinx ==4.3.2
- sphinx_rtd_theme ==1.0.0
- abspy *
- embreex ==2.17.7.post3
- sage-docbuild ==10.0
- sage-setup ==10.0
- sagemath-standard ==10.0



