NiaARM
NiaARM: A minimalistic framework for Numerical Association Rule Mining - Published in JOSS (2022)
Science Score: 98.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
✓CITATION.cff file
Found CITATION.cff file -
✓codemeta.json file
Found codemeta.json file -
✓.zenodo.json file
Found .zenodo.json file -
✓DOI references
Found 11 DOI reference(s) in README and JOSS metadata -
✓Academic publication links
Links to: arxiv.org, acm.org, joss.theoj.org -
○Committers with academic emails
-
○Institutional organization owner
-
✓JOSS paper metadata
Published in Journal of Open Source Software
Keywords
Keywords from Contributors
Scientific Fields
Repository
A minimalistic framework for Numerical Association Rule Mining
Basic Info
Statistics
- Stars: 19
- Watchers: 5
- Forks: 9
- Open Issues: 2
- Releases: 31
Topics
Metadata Files
README.md

NiaARM
A minimalistic framework for Numerical Association Rule Mining
🔍 Detailed insights • 📦 Installation • 🚀 Usage • 📄 Cite us • 📚 References • 📖 See also • 🔑 License • 🫂 Contributors
NiaARM is a framework for Association Rule Mining based on nature-inspired algorithms for optimization. 🌿 The framework is written fully in Python and runs on all platforms. NiaARM allows users to preprocess the data in a transaction database automatically, to search for association rules and provide a pretty output of the rules found. 📊 This framework also supports integral and real-valued types of attributes besides the categorical ones. Mining the association rules is defined as an optimization problem, and solved using the nature-inspired algorithms that come from the related framework called NiaPy. 🔗
- Documentation: https://niaarm.readthedocs.io/en/latest
- Tested OS: Windows, Ubuntu, Fedora, Alpine, Arch, macOS. However, that does not mean it does not work on others
🔍 Detailed insights
The current version includes (but is not limited to) the following functions:
- loading datasets in CSV format 📁
- preprocessing of data 🧹
- searching for association rules 🔎
- providing output of mined association rules 📋
- generating statistics about mined association rules 📊
- visualization of association rules 📈
- association rule text mining (experimental) 📄
📦 Installation
pip
To install NiaARM with pip, use:
sh
pip install niaarm
To install NiaARM on Alpine Linux, enable Community repository and use:
sh
$ apk add py3-niaarm
To install NiaARM on Arch Linux, use an AUR helper:
sh
$ yay -Syyu python-niaarm
To install NiaARM on Fedora, use:
sh
$ dnf install python3-niaarm
To install NiaARM on NixOS, use:
sh
nix-env -iA nixos.python311Packages.niaarm
🚀 Usage
Loading data
In NiaARM, data loading is done via the Dataset class. There are two options for loading data:
Option 1: From a pandas DataFrame (recommended)
```python import pandas as pd from niaarm import Dataset
df = pd.read_csv('datasets/Abalone.csv')
preprocess data...
data = Dataset(df) print(data) # printing the dataset will generate a feature report ```
Option 2: Directly from a CSV file
```python from niaarm import Dataset
data = Dataset('datasets/Abalone.csv') print(data) ```
Preprocessing
Data Squashing
Optionally, a preprocessing technique, called data squashing [5], can be applied. This will significantly reduce the number of transactions, while providing similar results to the original dataset.
```python from niaarm import Dataset, squash
dataset = Dataset('datasets/Abalone.csv') squashed = squash(dataset, threshold=0.9, similarity='euclidean') print(squashed) ```
Mining association rules
The easy way (recommended)
Association rule mining can be easily performed using the get_rules function:
```python from niaarm import Dataset, get_rules from niapy.algorithms.basic import DifferentialEvolution
data = Dataset("datasets/Abalone.csv")
algo = DifferentialEvolution(populationsize=50, differentialweight=0.5, crossover_probability=0.9) metrics = ('support', 'confidence')
rules, runtime = getrules(data, algo, metrics, max_iters=30, logging=True)
print(rules) # Prints basic stats about the mined rules print(f'Run Time: {runtime}') rules.tocsv('output.csv') ```
The hard way
The above example can be also be implemented using a more low level interface,
with the NiaARM class directly:
```python from niaarm import NiaARM, Dataset from niapy.algorithms.basic import DifferentialEvolution from niapy.task import Task, OptimizationType
data = Dataset("datasets/Abalone.csv")
Create a problem
dimension represents the dimension of the problem;
features represent the list of features, while transactions depicts the list of transactions
metrics is a sequence of metrics to be taken into account when computing the fitness;
you can also pass in a dict of the shape {'metric_name': };
when passing a sequence, the weights default to 1.
problem = NiaARM(data.dimension, data.features, data.transactions, metrics=('support', 'confidence'), logging=True)
build niapy task
task = Task(problem=problem, maxiters=30, optimizationtype=OptimizationType.MAXIMIZATION)
use Differential Evolution (DE) algorithm from the NiaPy library
see full list of available algorithms: https://github.com/NiaOrg/NiaPy/blob/master/Algorithms.md
algo = DifferentialEvolution(populationsize=50, differentialweight=0.5, crossover_probability=0.9)
run algorithm
best = algo.run(task=task)
sort rules
problem.rules.sort()
export all rules to csv
problem.rules.to_csv('output.csv') ```
Interest measures
The framework implements several popular interest measures, which can be used to compute the fitness function value of rules and for assessing the quality of the mined rules. A full list of the implemented interest measures along with their descriptions and equations can be found here.
Visualization
The framework currently supports (visualizations):
- hill slopes (presented in [4]),
- scatter plot and
- grouped matrix plot visualization methods.
More visualization methods are planned to be implemented in future releases.
Hill Slopes
```python from matplotlib import pyplot as plt from niaarm import Dataset, getrules from niaarm.visualize import hillslopes
dataset = Dataset('datasets/Abalone.csv') metrics = ('support', 'confidence') rules, _ = getrules(dataset, 'DifferentialEvolution', metrics, maxevals=1000, seed=1234) somerule = rules[150] hillslopes(some_rule, dataset.transactions) plt.show() ```
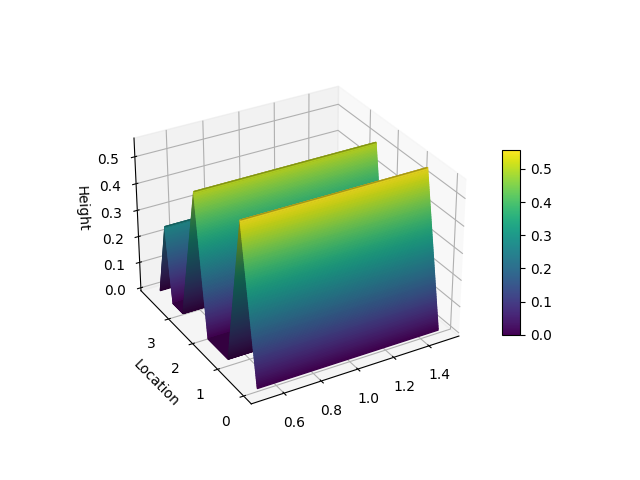
Scatter Plot
```python from examples.visualizationexamples.preparedatasets import getweatherdata from niaarm import Dataset, getrules from niaarm.visualize import scatterplot
Get prepared data
armdf = getweather_data()
Prepare Dataset
dataset = Dataset(pathordf=arm_df,delimiter=",")
Get rules
metrics = ("support", "confidence") rules, runtime = getrules(dataset, "DifferentialEvolution", metrics, max_evals=500)
Add lift to metrics
metrics = list(metrics) metrics.append("lift") metrics = tuple(metrics)
Visualize scatter plot
fig = scatter_plot(rules=rules, metrics=metrics, interactive=False) fig.show() ```

Grouped Matrix Plot
```python from examples.visualizationexamples.preparedatasets import getfootballplayerdata from niaarm import Dataset, getrules from niaarm.visualize import groupedmatrixplot
Get prepared data
armdf = getfootballplayerdata()
Prepare Dataset
dataset = Dataset(pathordf=arm_df, delimiter=",")
Get rules
metrics = ("support", "confidence") rules, runtime = getrules(dataset, "DifferentialEvolution", metrics, max_evals=500)
Add lift to metrics
metrics = list(metrics) metrics.append("lift") metrics = tuple(metrics)
Visualize grouped matrix plot
fig = groupedmatrixplot(rules=rules, metrics=metrics, k=5, interactive=False) fig.show() ```

Text Mining (Experimental)
An experimental implementation of association rule text mining using nature-inspired algorithms, based on ideas from [5]
is also provided. The niaarm.text module contains the Corpus and Document classes for loading and preprocessing corpora,
a TextRule class, representing a text rule, and the NiaARTM class, implementing association rule text mining
as a continuous optimization problem. The get_text_rules function, equivalent to get_rules, but for text mining, was also
added to the niaarm.mine module.
```python import pandas as pd from niaarm.text import Corpus from niaarm.mine import gettextrules from niapy.algorithms.basic import ParticleSwarmOptimization
df = pd.readjson('datasets/text/artmtestdataset.json', orient='records') documents = df['text'].tolist() corpus = Corpus.fromlist(documents)
algorithm = ParticleSwarmOptimization(populationsize=200, seed=123) metrics = ('support', 'confidence', 'aws') rules, time = gettextrules(corpus, maxterms=5, algorithm=algorithm, metrics=metrics, max_evals=10000, logging=True)
print(rules) print(f'Run time: {time:.2f}s') rules.to_csv('output.csv') ```
Note: You may need to download stopwords and the punkt tokenizer from nltk by running import nltk; nltk.download('stopwords'); nltk.download('punkt').
For a full list of examples see the examples folder in the GitHub repository.
Command line interface
We provide a simple command line interface, which allows you to easily mine association rules on any input dataset, output them to a csv file and/or perform a simple statistical analysis on them. For more details see the documentation.
shell
niaarm -h
``` usage: niaarm [-h] [-v] [-c CONFIG] [-i INPUT_FILE] [-o OUTPUT_FILE] [--squashing-similarity {euclidean,cosine}] [--squashing-threshold SQUASHING_THRESHOLD] [-a ALGORITHM] [-s SEED] [--max-evals MAX_EVALS] [--max-iters MAX_ITERS] [--metrics METRICS [METRICS ...]] [--weights WEIGHTS [WEIGHTS ...]] [--log] [--stats]
Perform ARM, output mined rules as csv, get mined rules' statistics
options:
-h, --help show this help message and exit
-v, --version show program's version number and exit
-c CONFIG, --config CONFIG
Path to a TOML config file
-i INPUTFILE, --input-file INPUTFILE
Input file containing a csv dataset
-o OUTPUTFILE, --output-file OUTPUTFILE
Output file for mined rules
--squashing-similarity {euclidean,cosine}
Similarity measure to use for squashing
--squashing-threshold SQUASHINGTHRESHOLD
Threshold to use for squashing
-a ALGORITHM, --algorithm ALGORITHM
Algorithm to use (niapy class name, e.g. DifferentialEvolution)
-s SEED, --seed SEED Seed for the algorithm's random number generator
--max-evals MAXEVALS
Maximum number of fitness function evaluations
--max-iters MAX_ITERS
Maximum number of iterations
--metrics METRICS [METRICS ...]
Metrics to use in the fitness function.
--weights WEIGHTS [WEIGHTS ...]
Weights in range [0, 1] corresponding to --metrics
--log Enable logging of fitness improvements
--stats Display stats about mined rules
``
Note: The CLI script can also run as a python module (python -m niaarm ...`)
📄 Cite us
Stupan, Ž., & Fister Jr., I. (2022). NiaARM: A minimalistic framework for Numerical Association Rule Mining. Journal of Open Source Software, 7(77), 4448.
📚 References
Ideas are based on the following research papers:
[1] I. Fister Jr., A. Iglesias, A. Gálvez, J. Del Ser, E. Osaba, I Fister. Differential evolution for association rule mining using categorical and numerical attributes In: Intelligent data engineering and automated learning - IDEAL 2018, pp. 79-88, 2018.
[2] I. Fister Jr., V. Podgorelec, I. Fister. Improved Nature-Inspired Algorithms for Numeric Association Rule Mining. In: Vasant P., Zelinka I., Weber GW. (eds) Intelligent Computing and Optimization. ICO 2020. Advances in Intelligent Systems and Computing, vol 1324. Springer, Cham.
[3] I. Fister Jr., I. Fister A brief overview of swarm intelligence-based algorithms for numerical association rule mining. arXiv preprint arXiv:2010.15524 (2020).
[4] Fister, I. et al. (2020). Visualization of Numerical Association Rules by Hill Slopes. In: Analide, C., Novais, P., Camacho, D., Yin, H. (eds) Intelligent Data Engineering and Automated Learning – IDEAL 2020. IDEAL 2020. Lecture Notes in Computer Science(), vol 12489. Springer, Cham. https://doi.org/10.1007/978-3-030-62362-3_10
[5] I. Fister, S. Deb, I. Fister, Population-based metaheuristics for Association Rule Text Mining, In: Proceedings of the 2020 4th International Conference on Intelligent Systems, Metaheuristics & Swarm Intelligence, New York, NY, USA, mar. 2020, pp. 19–23. doi: 10.1145/3396474.3396493.
[6] I. Fister, I. Fister Jr., D. Novak and D. Verber, Data squashing as preprocessing in association rule mining, 2022 IEEE Symposium Series on Computational Intelligence (SSCI), Singapore, Singapore, 2022, pp. 1720-1725, doi: 10.1109/SSCI51031.2022.10022240.
📖 See also
[1] NiaARM.jl: Numerical Association Rule Mining in Julia
🔑 License
This package is distributed under the MIT License. This license can be found online at http://www.opensource.org/licenses/MIT.
Disclaimer
This framework is provided as-is, and there are no guarantees that it fits your purposes or that it is bug-free. Use it at your own risk!
🫂 Contributors
Thanks goes to these wonderful people (emoji key):
zStupan 💻 🐛 📖 🖋 🤔 💡 |
Iztok Fister Jr. 💻 🐛 🧑🏫 🚧 🤔 |
Erkan Karabulut 💻 🐛 |
Tadej Lahovnik 📖 |
Ben Beasley 📖 |
Dusan Fister 🎨 |
This project follows the all-contributors specification. Contributions of any kind welcome!
Owner
- Name: Iztok Fister Jr.
- Login: firefly-cpp
- Kind: user
- Location: Slovenia
- Website: http://www.iztok-jr-fister.eu/
- Repositories: 28
- Profile: https://github.com/firefly-cpp
JOSS Publication
NiaARM: A minimalistic framework for Numerical Association Rule Mining
Authors
Tags
association rule mining data mining evolutionary algorithms numerical association rule mining visualizationCitation (CITATION.cff)
cff-version: 1.2.0 message: "If you use this software, please cite it as below." authors: - family-names: "Stupan" given-names: "Žiga" orcid: "https://orcid.org/0000-0001-9847-7306" - family-names: "Fister Jr." given-names: "Iztok" orcid: "https://orcid.org/0000-0002-6418-1272" title: "NiaARM: A minimalistic framework for Numerical Association Rule Mining" version: 0.2.1 doi: 10.21105/joss.04448 date-released: 2022-09-29 url: "https://github.com/firefly-cpp/NiaARM"
GitHub Events
Total
- Create event: 5
- Issues event: 4
- Release event: 5
- Watch event: 3
- Delete event: 3
- Issue comment event: 7
- Member event: 1
- Push event: 18
- Pull request event: 24
- Fork event: 7
Last Year
- Create event: 5
- Issues event: 4
- Release event: 5
- Watch event: 3
- Delete event: 3
- Issue comment event: 7
- Member event: 1
- Push event: 18
- Pull request event: 24
- Fork event: 7
Committers
Last synced: 7 months ago
Top Committers
| Name | Commits | |
|---|---|---|
| zStupan | z****n@g****m | 173 |
| firefly-cpp | i****k@i****u | 139 |
| dependabot[bot] | 4****] | 24 |
| allcontributors[bot] | 4****] | 12 |
| Miha Bukovnik | m****k@p****i | 9 |
| Tadej Lahovnik | t****k@s****i | 8 |
| Zan Vrabic | z****c@s****i | 7 |
| rhododendrom | b****2@p****m | 6 |
| HlisTilen | 4****n | 4 |
| howsun.jow | h****w@g****m | 3 |
| Miha Bukovnik | z****k@g****m | 2 |
| erkankarabulut | e****t@g****m | 2 |
| Arfon Smith | a****n@g****m | 1 |
| Benjamin A. Beasley | c****e@m****t | 1 |
Committer Domains (Top 20 + Academic)
Issues and Pull Requests
Last synced: 6 months ago
All Time
- Total issues: 56
- Total pull requests: 112
- Average time to close issues: 3 months
- Average time to close pull requests: 1 day
- Total issue authors: 10
- Total pull request authors: 12
- Average comments per issue: 1.82
- Average comments per pull request: 0.43
- Merged pull requests: 104
- Bot issues: 0
- Bot pull requests: 37
Past Year
- Issues: 4
- Pull requests: 26
- Average time to close issues: 5 months
- Average time to close pull requests: 3 days
- Issue authors: 2
- Pull request authors: 5
- Average comments per issue: 1.25
- Average comments per pull request: 0.35
- Merged pull requests: 22
- Bot issues: 0
- Bot pull requests: 7
Top Authors
Issue Authors
- firefly-cpp (42)
- zStupan (3)
- erkankarabulut (2)
- howsunjow (2)
- RamaSubramanianT (1)
- minakshikaushik (1)
- mlaky88 (1)
- carlosal1015 (1)
- BukovnikMiha (1)
- fabian-s (1)
Pull Request Authors
- zStupan (49)
- dependabot[bot] (47)
- allcontributors[bot] (11)
- lahovniktadej (10)
- vrabiczan (6)
- erkankarabulut (4)
- HlisTilen (4)
- BukovnikMiha (3)
- howsunjow (3)
- firefly-cpp (3)
- arfon (1)
- musicinmybrain (1)
Top Labels
Issue Labels
Pull Request Labels
Packages
- Total packages: 21
-
Total downloads:
- pypi 305 last-month
-
Total dependent packages: 3
(may contain duplicates) -
Total dependent repositories: 1
(may contain duplicates) - Total versions: 119
- Total maintainers: 2
alpine-v3.18: py3-niaarm-pyc
Precompiled Python bytecode for py3-niaarm
- Homepage: https://github.com/firefly-cpp/NiaARM
- License: MIT
-
Latest release: 0.3.1-r1
published almost 3 years ago
Rankings
Maintainers (1)
alpine-v3.18: py3-niaarm
A minimalistic framework for numerical association rule mining
- Homepage: https://github.com/firefly-cpp/NiaARM
- License: MIT
-
Latest release: 0.3.1-r1
published almost 3 years ago
Rankings
Maintainers (1)
alpine-v3.18: py3-niaarm-doc
A minimalistic framework for numerical association rule mining (documentation)
- Homepage: https://github.com/firefly-cpp/NiaARM
- License: MIT
-
Latest release: 0.3.1-r1
published almost 3 years ago
Rankings
Maintainers (1)
pypi.org: niaarm
A minimalistic framework for numerical association rule mining
- Homepage: https://github.com/firefly-cpp/NiaARM
- Documentation: https://niaarm.readthedocs.io/en/latest/
- License: MIT
-
Latest release: 0.4.3
published 7 months ago
Rankings
Maintainers (1)
alpine-edge: py3-niaarm-doc
A minimalistic framework for numerical association rule mining (documentation)
- Homepage: https://github.com/firefly-cpp/NiaARM
- License: MIT AND Apache-2.0
-
Latest release: 0.4.3-r0
published 7 months ago
Rankings
Maintainers (1)
alpine-edge: py3-niaarm
A minimalistic framework for numerical association rule mining
- Homepage: https://github.com/firefly-cpp/NiaARM
- License: MIT AND Apache-2.0
-
Latest release: 0.4.3-r0
published 7 months ago
Rankings
Maintainers (1)
alpine-edge: py3-niaarm-pyc
Precompiled Python bytecode for py3-niaarm
- Homepage: https://github.com/firefly-cpp/NiaARM
- License: MIT AND Apache-2.0
-
Latest release: 0.4.3-r0
published 7 months ago
Rankings
Maintainers (1)
alpine-v3.17: py3-niaarm
A minimalistic framework for numerical association rule mining
- Homepage: https://github.com/firefly-cpp/NiaARM
- License: MIT
-
Latest release: 0.2.2-r3
published over 3 years ago
Rankings
Maintainers (1)
alpine-v3.17: py3-niaarm-doc
A minimalistic framework for numerical association rule mining (documentation)
- Homepage: https://github.com/firefly-cpp/NiaARM
- License: MIT
-
Latest release: 0.2.2-r3
published over 3 years ago
Rankings
Maintainers (1)
alpine-v3.22: py3-niaarm-pyc
Precompiled Python bytecode for py3-niaarm
- Homepage: https://github.com/firefly-cpp/NiaARM
- License: MIT AND Apache-2.0
-
Latest release: 0.4.1-r0
published 12 months ago
Rankings
Maintainers (1)
alpine-v3.19: py3-niaarm-pyc
Precompiled Python bytecode for py3-niaarm
- Homepage: https://github.com/firefly-cpp/NiaARM
- License: MIT
-
Latest release: 0.3.5-r0
published over 2 years ago
Rankings
Maintainers (1)
alpine-v3.20: py3-niaarm
A minimalistic framework for numerical association rule mining
- Homepage: https://github.com/firefly-cpp/NiaARM
- License: MIT
-
Latest release: 0.3.9-r0
published almost 2 years ago
Rankings
Maintainers (1)
alpine-v3.21: py3-niaarm-doc
A minimalistic framework for numerical association rule mining (documentation)
- Homepage: https://github.com/firefly-cpp/NiaARM
- License: MIT AND Apache-2.0
-
Latest release: 0.3.12-r1
published over 1 year ago
Rankings
Maintainers (1)
alpine-v3.19: py3-niaarm-doc
A minimalistic framework for numerical association rule mining (documentation)
- Homepage: https://github.com/firefly-cpp/NiaARM
- License: MIT
-
Latest release: 0.3.5-r0
published over 2 years ago
Rankings
alpine-v3.22: py3-niaarm-doc
A minimalistic framework for numerical association rule mining (documentation)
- Homepage: https://github.com/firefly-cpp/NiaARM
- License: MIT AND Apache-2.0
-
Latest release: 0.4.1-r0
published 12 months ago
Rankings
Maintainers (1)
alpine-v3.20: py3-niaarm-doc
A minimalistic framework for numerical association rule mining (documentation)
- Homepage: https://github.com/firefly-cpp/NiaARM
- License: MIT
-
Latest release: 0.3.9-r0
published almost 2 years ago
Rankings
Maintainers (1)
alpine-v3.19: py3-niaarm
A minimalistic framework for numerical association rule mining
- Homepage: https://github.com/firefly-cpp/NiaARM
- License: MIT
-
Latest release: 0.3.5-r0
published over 2 years ago
Rankings
Maintainers (1)
alpine-v3.20: py3-niaarm-pyc
Precompiled Python bytecode for py3-niaarm
- Homepage: https://github.com/firefly-cpp/NiaARM
- License: MIT
-
Latest release: 0.3.9-r0
published almost 2 years ago
Rankings
Maintainers (1)
alpine-v3.21: py3-niaarm
A minimalistic framework for numerical association rule mining
- Homepage: https://github.com/firefly-cpp/NiaARM
- License: MIT AND Apache-2.0
-
Latest release: 0.3.12-r1
published over 1 year ago
Rankings
Maintainers (1)
alpine-v3.21: py3-niaarm-pyc
Precompiled Python bytecode for py3-niaarm
- Homepage: https://github.com/firefly-cpp/NiaARM
- License: MIT AND Apache-2.0
-
Latest release: 0.3.12-r1
published over 1 year ago
Rankings
Maintainers (1)
alpine-v3.22: py3-niaarm
A minimalistic framework for numerical association rule mining
- Homepage: https://github.com/firefly-cpp/NiaARM
- License: MIT AND Apache-2.0
-
Latest release: 0.4.1-r0
published 12 months ago
Rankings
Maintainers (1)
Dependencies
- Sphinx ==4.4.0
- niapy >=2.0.1
- numpy >=1.22.3
- pandas >=1.4.0
- sphinx-rtd-theme ==1.0.0
- sphinxcontrib-bibtex ==2.4.1
- atomicwrites 1.4.0 develop
- attrs 21.4.0 develop
- iniconfig 1.1.1 develop
- pluggy 1.0.0 develop
- py 1.11.0 develop
- pytest 7.1.2 develop
- alabaster 0.7.12
- babel 2.10.1
- certifi 2022.5.18.1
- charset-normalizer 2.0.12
- click 8.1.3
- colorama 0.4.4
- cycler 0.11.0
- docutils 0.17.1
- et-xmlfile 1.1.0
- fonttools 4.33.3
- idna 3.3
- imagesize 1.3.0
- importlib-metadata 4.11.4
- jinja2 3.1.2
- joblib 1.1.0
- kiwisolver 1.4.2
- latexcodec 2.0.1
- markupsafe 2.1.1
- matplotlib 3.5.2
- niapy 2.0.2
- nltk 3.7
- numpy 1.21.6
- numpy 1.22.4
- openpyxl 3.0.10
- packaging 21.3
- pandas 1.3.5
- pandas 1.4.2
- pillow 9.1.1
- pybtex 0.24.0
- pybtex-docutils 1.0.1
- pygments 2.12.0
- pyparsing 3.0.9
- python-dateutil 2.8.2
- pytz 2022.1
- pyyaml 6.0
- regex 2022.4.24
- requests 2.27.1
- setuptools-scm 6.4.2
- six 1.16.0
- snowballstemmer 2.2.0
- sphinx 4.5.0
- sphinx-rtd-theme 1.0.0
- sphinxcontrib-applehelp 1.0.2
- sphinxcontrib-bibtex 2.4.2
- sphinxcontrib-devhelp 1.0.2
- sphinxcontrib-htmlhelp 2.0.0
- sphinxcontrib-jsmath 1.0.1
- sphinxcontrib-qthelp 1.0.3
- sphinxcontrib-serializinghtml 1.1.5
- tomli 2.0.1
- tqdm 4.64.0
- typing-extensions 4.2.0
- urllib3 1.26.9
- zipp 3.8.0
- pytest ^7.0.1 develop
- Sphinx ^4.4.0
- niapy ^2.0.1
- nltk ^3.7
- numpy --- - !ruby/hash:ActiveSupport::HashWithIndifferentAccess version: "^1.21.5" python: ">=3.7,<3.11" - !ruby/hash:ActiveSupport::HashWithIndifferentAccess version: "^1.22.3" python: "^3.11"
- pandas --- - !ruby/hash:ActiveSupport::HashWithIndifferentAccess version: "^1.3.5" python: ">=3.7.1,<3.8" - !ruby/hash:ActiveSupport::HashWithIndifferentAccess version: "^1.4.0" python: "^3.8"
- python ^3.7
- sphinx-rtd-theme ^1.0.0
- sphinxcontrib-bibtex ^2.4.1
- actions/cache v3 composite
- actions/checkout v3 composite
- actions/setup-python v3 composite








