https://github.com/bluegreen-labs/skytrackr
A Sky Illuminance Location Tracker
Science Score: 36.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
○CITATION.cff file
-
✓codemeta.json file
Found codemeta.json file -
○.zenodo.json file
-
✓DOI references
Found 5 DOI reference(s) in README -
✓Academic publication links
Links to: zenodo.org -
○Committers with academic emails
-
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (16.7%) to scientific vocabulary
Keywords
Repository
A Sky Illuminance Location Tracker
Basic Info
- Host: GitHub
- Owner: bluegreen-labs
- License: agpl-3.0
- Language: R
- Default Branch: main
- Homepage: https://bluegreen-labs.github.io/skytrackr/
- Size: 4.42 MB
Statistics
- Stars: 0
- Watchers: 0
- Forks: 0
- Open Issues: 2
- Releases: 1
Topics
Metadata Files
README.md
skytrackr
A tool to calculate geolocation by light positions using sky illuminance values (in lux). The routine uses model optimization and parameter estimation to determine locations robustly.
How to cite this package
You can cite this package like this "we obtained location estimates using the {skytrackr} R package (Hufkens 2023)". Here is the full bibliographic reference to include in your reference list (don't forget to update the 'last accessed' date):
Hufkens, K. (2023). skytrackr: a sky illuminance location tracker. Zenodo. https://doi.org/10.5281/zenodo.8331492.
Installation
development release
To install the development releases of the package run the following commands:
r
if(!require(remotes)){install.packages("remotes")}
remotes::install_github("bluegreen-labs/skytrackr")
library("skytrackr")
Vignettes are not rendered by default, if you want to include additional documentation please use:
r
if(!require(remotes)){install.packages("remotes")}
remotes::install_github("bluegreen-labs/skytrackr", build_vignettes = TRUE)
library("skytrackr")
Use
To demonstrate the functioning of the package a small demo dataset comprised of a couple of days of light logging of a Common swift near a nest box in Ghent, Belgium was included (i.e. tag cc876). I will use this data to demonstrate the features of the package. Note that when multiple dates are present all dates will be considered. The package is friendly to the use of R piped (|>) commands. Using all default settings you can just pipe the data in to the skytrackr() function. The returned object will be a data frame containing the best estimate (median) of the longitude and latitude as well as 5-95% quantile as sampled from the posterior parameter distribution.
Data pre-screening
During the breeding season it is advised to remove data which is affected by time spent on a nest, altering the light signal and position estimates. Both the light patterns and other sensory data can be used to restrict the data to migration and non-breeding movements.
In the package I include the stk_profile() function which plots .lux files (and their ancillary .deg data) in an overview plot. The default plot is a static plot, however calling the function using the plotly = TRUE argument will render a dynamic plot on which you can zoom and which provides a tooltip showing the data values (including the date / time).
```r
read in the lux file, if a matching .deg
file is found it is integrated into one
long oriented data format
(here the internal demo data is used)
df <- stkreadlux( system.file("extdata/cc876.lux", package="skytrackr") )
plot the data as a static plot
stk_profile(df)
plot the data using plotly
(this allows exploring data values, e.g. zooming in and using a tooltip)
stk_profile(df, plotly = TRUE) ```
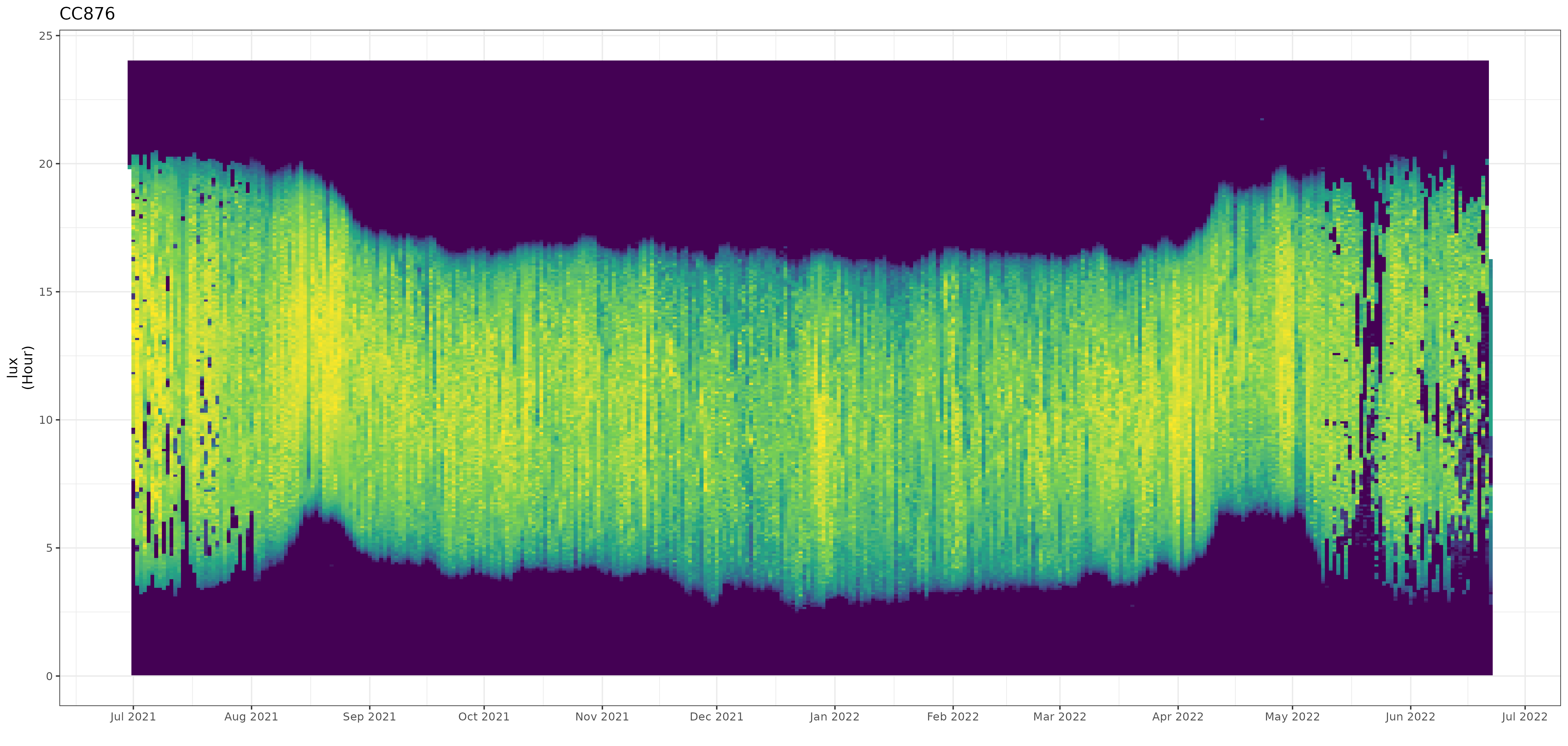
Date and time values of the non-breeding season can easily be determined and used in further sub-setting of the data for final processing. You can use the automated stk_screen_twl() function to remove dates with either stationary roosting behaviour (with dark values during daytime hours), or with "false" twilight events (due to birds roosting in dark conditions).
```r
filter data using twilight heuristics
and filter out the flagged values
df <- df |> stkscreentwl(filter = TRUE)
you can run the stk_profile() command again
to show the trimmed data, note all functions
are pipeable
df |> stk_profile() ```
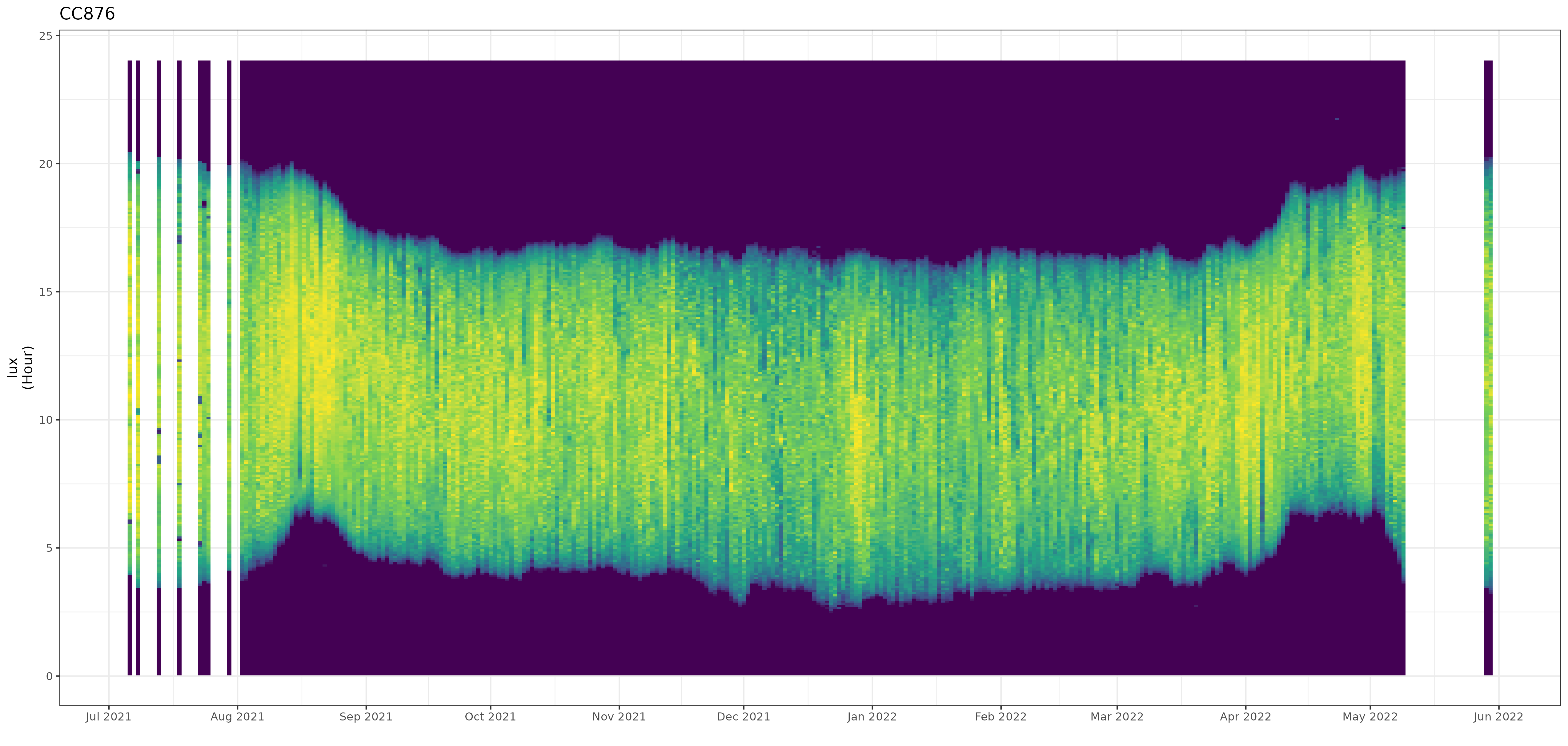
Further sub-setting can be done from this point, but if a bird is known to remain in a single location for breeding one can assume the individual is stationary. The filtering is there to provide as much of an hands off approach as possible, but use expert judgement.
MCMC DEzs optimization
To track movements by light using {skytrackr} you will need a few additional parameters. First, you will need to define the methodology used to find an optimal solution. The underlying BayesianTools package used in optimization allows for the specification of optimization techniques using the 'control' statement. The same argument can be used in the main skytrackr() routine.
In addition, care should be taken to specify the start location and tolerance (maximum degrees covered in a single flight - in km). The routine also needs you to specify an applicable land mask for land bound species. This land mask can be buffered extending somewhat offshore if desired. Finally, there is a behavioural component to the optimization routine used through the use of a step-selection function. This step-selection function determines the probability that a certain distance is covered based upon a probability density distribution provided by the user. Common distributions within movement ecology are for example a gamma or weibull distribution. Ideally these distributions are fit to previously collected data (either light-logger or GPS based), or based on common sense.
All three factors, the tolerance (maximum distance covered), the land mask (limiting locations to those over land), and step-selection function (providing a probabilistic constrained on distance covered), constrain the parameter fit. This ensures stability of results on a pseudo-mechanistic basis.
```r
define land mask with a bounding box
and an off-shore buffer (in km), in addition
you can specifiy the resolution of the resulting raster
mask <- stk_mask( bbox = c(-20, -40, 60, 60), #xmin, ymin, xmax, ymax buffer = 150, # in km resolution = 0.5 )
define a step selection distribution
ssf <- function(x, shape = 1.02, scale = 250, tolerance = 1500){ norm <- sum(stats::dgamma(1:tolerance, shape = shape, scale = scale)) prob <- stats::dgamma(x, shape = shape, scale = scale) / norm }
locations <- data |> skytrackr( startlocation = c(51.08, 3.73), tolerance = 1500, # in km scale = c(1,10), range = c(1.5, 140), control = list( sampler = 'DEzs', settings = list( burnin = 500, iterations = 1500, message = FALSE ) ), stepselection = ssf, mask = mask, plot = TRUE ) ```
If you enable plotting during the optimization routine a plot will be drawn with each new location which is determined. The plot shows a map covering the geographic extent as set by the mask you provide and a green region of interest defined by the intersection of the mask and a hard distance threshold tolerance (tolerance parameter). The start position is indicated with a black triangle, the latest position is defined by a closed and open circle combined.
During equinoxes the small closed circles will be small open circles as these periods have increased inherent uncertainty. To the right are also three panels showing the best value of the three estimated parameters as a black line, and their uncertainty intervals. Note that using the plotting option considerably slows down overall calculation due to rendering times (it is fun to watch though).
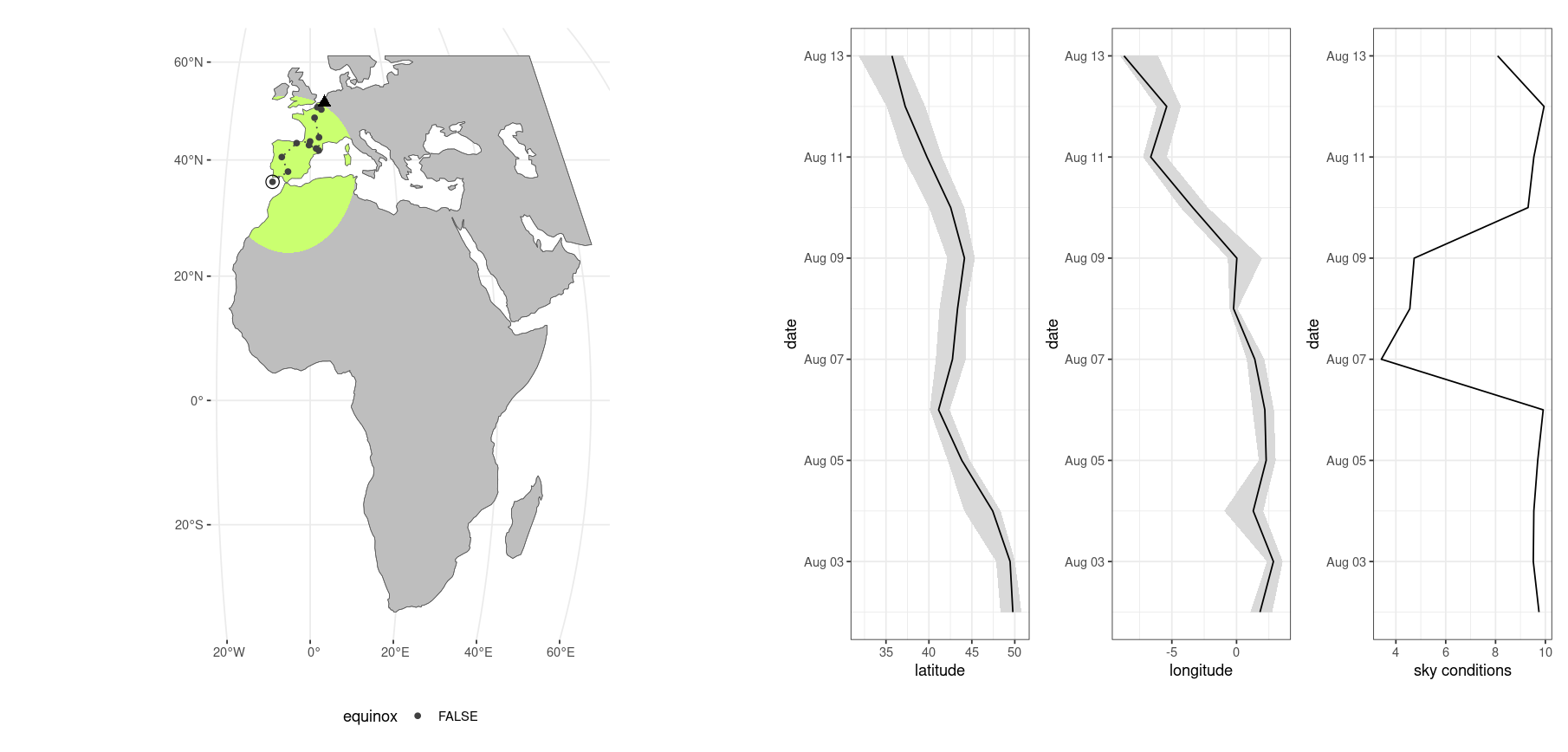
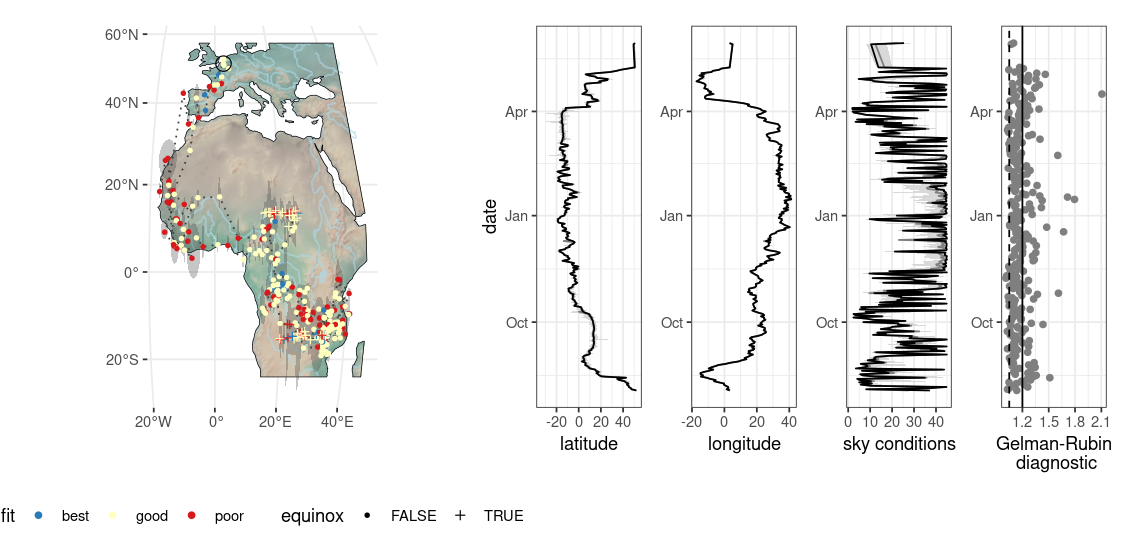
You can map the final location estimates using the same rendering layout using stk_map().
r
locations |> stk_map(bbox = c(-20, -40, 60, 60))
Owner
- Name: BlueGreen Labs
- Login: bluegreen-labs
- Kind: organization
- Email: info@bluegreenlabs.org
- Location: Melsele, Belgium
- Website: http://bluegreenlabs.org
- Repositories: 17
- Profile: https://github.com/bluegreen-labs
BlueGreen open science labs & consulting, providing environmental research infrastructure and editorial solutions.
GitHub Events
Total
- Push event: 4
Last Year
- Push event: 4
Committers
Last synced: about 2 years ago
Top Committers
| Name | Commits | |
|---|---|---|
| Koen Hufkens | k****s@g****m | 109 |
Issues and Pull Requests
Last synced: 11 months ago
All Time
- Total issues: 7
- Total pull requests: 6
- Average time to close issues: 8 days
- Average time to close pull requests: about 4 hours
- Total issue authors: 1
- Total pull request authors: 1
- Average comments per issue: 0.43
- Average comments per pull request: 0.17
- Merged pull requests: 6
- Bot issues: 0
- Bot pull requests: 0
Past Year
- Issues: 3
- Pull requests: 0
- Average time to close issues: 2 days
- Average time to close pull requests: N/A
- Issue authors: 1
- Pull request authors: 0
- Average comments per issue: 0.0
- Average comments per pull request: 0
- Merged pull requests: 0
- Bot issues: 0
- Bot pull requests: 0
Top Authors
Issue Authors
- khufkens (6)
Pull Request Authors
- khufkens (6)
Top Labels
Issue Labels
Pull Request Labels
Dependencies
- actions/checkout v3 composite
- r-lib/actions/check-r-package v2 composite
- r-lib/actions/setup-pandoc v2 composite
- r-lib/actions/setup-r v2 composite
- r-lib/actions/setup-r-dependencies v2 composite
- JamesIves/github-pages-deploy-action v4.4.1 composite
- actions/checkout v3 composite
- r-lib/actions/setup-pandoc v2 composite
- r-lib/actions/setup-r v2 composite
- r-lib/actions/setup-r-dependencies v2 composite
- actions/checkout v2 composite
- r-lib/actions/setup-pandoc v2 composite
- r-lib/actions/setup-r v2 composite
- R >= 4.2 depends
- BayesianTools * imports
- graphics * imports
- maps * imports
- progress * imports
- skylight * imports
- stats * imports
- utils * imports
- covr * suggests
- dplyr * suggests
- ggplot2 * suggests
- knitr * suggests
- patchwork * suggests
- rmarkdown * suggests
- rnaturalearth * suggests
- testthat * suggests