https://github.com/brainglobe/cellfinder-napari
Efficient cell detection in large images using cellfinder in napari
Science Score: 23.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
○CITATION.cff file
-
○codemeta.json file
-
○.zenodo.json file
-
✓DOI references
Found 6 DOI reference(s) in README -
○Academic publication links
-
✓Committers with academic emails
2 of 9 committers (22.2%) from academic institutions -
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (14.4%) to scientific vocabulary
Keywords
Keywords from Contributors
Repository
Efficient cell detection in large images using cellfinder in napari
Basic Info
- Host: GitHub
- Owner: brainglobe
- License: bsd-3-clause
- Language: Python
- Default Branch: main
- Homepage: https://brainglobe.info/documentation/cellfinder/index.html
- Size: 34.3 MB
Statistics
- Stars: 22
- Watchers: 3
- Forks: 6
- Open Issues: 0
- Releases: 0
Topics
Metadata Files
README.md
This package has moved
cellfinder-napari has merged with it's backend code and is now available as a single package called cellfinder.
We recommend you uninstall cellfinder-napari and instead use the functionality provided in the cellfinder package.
These changes are part of our wider restructuring of the BrainGlobe suite of tools and analysis pipelines, which you can keep up to date with on our blog.
cellfinder-napari
Efficient cell detection in large images (e.g. whole mouse brain images)
cellfinder-napari is a front-end to cellfinder-core to allow ease of use within the napari multidimensional image viewer. For more details on this approach, please see Tyson, Rousseau & Niedworok et al. (2021). This algorithm can also be used within the original
cellfinder software for
whole-brain microscopy analysis.
cellfinder-napari, cellfinder and cellfinder-core were developed by Charly Rousseau and Adam Tyson in the Margrie Lab, based on previous work by Christian Niedworok, generously supported by the Sainsbury Wellcome Centre.
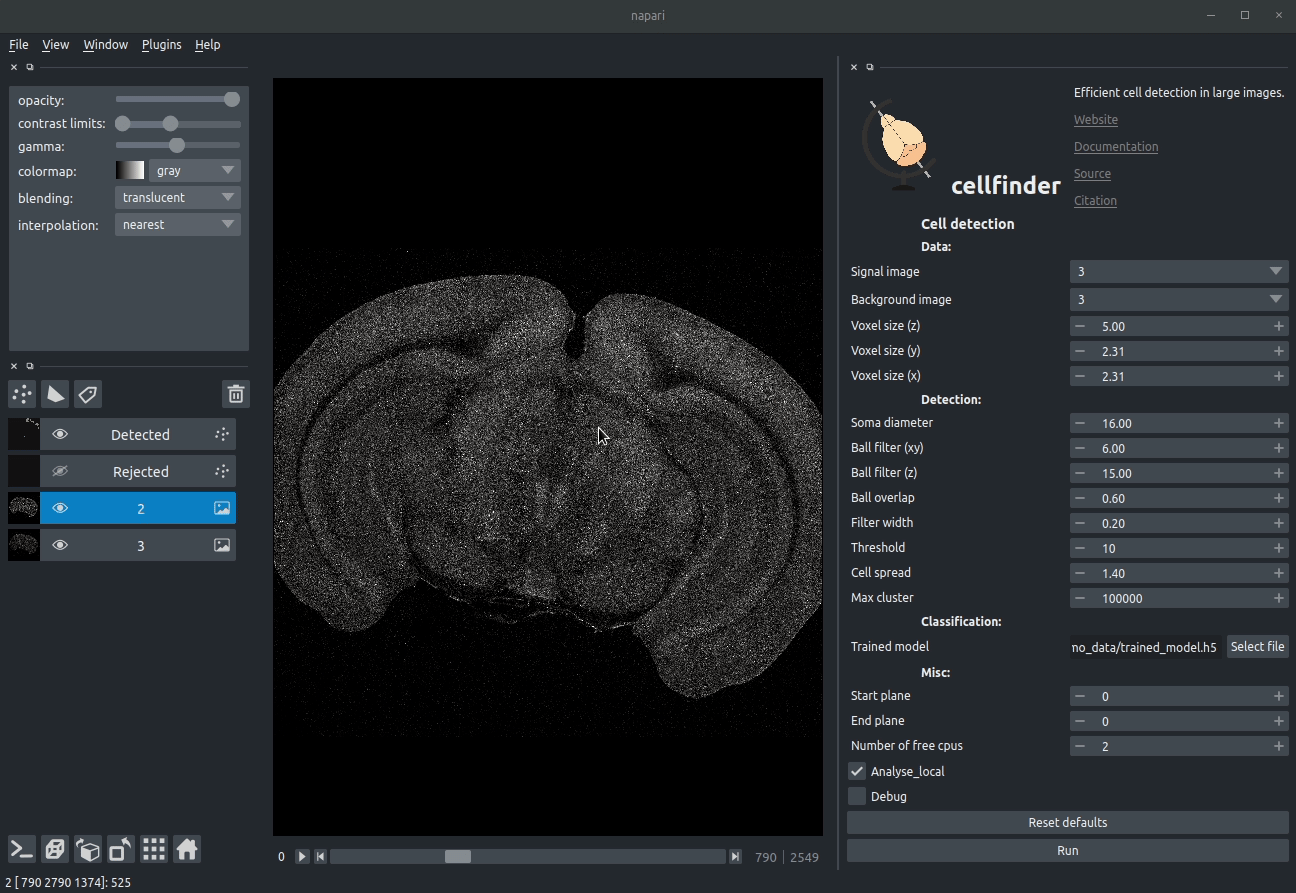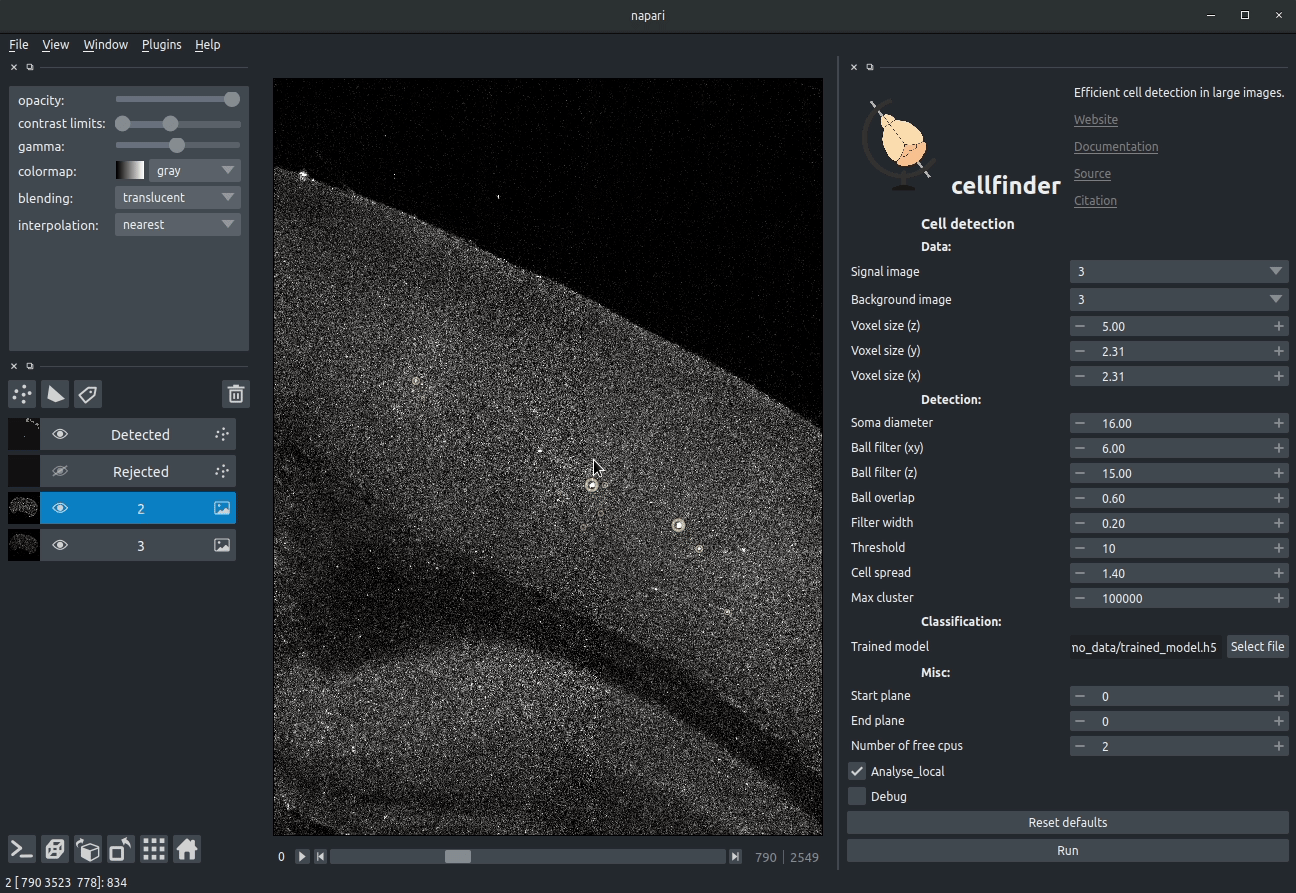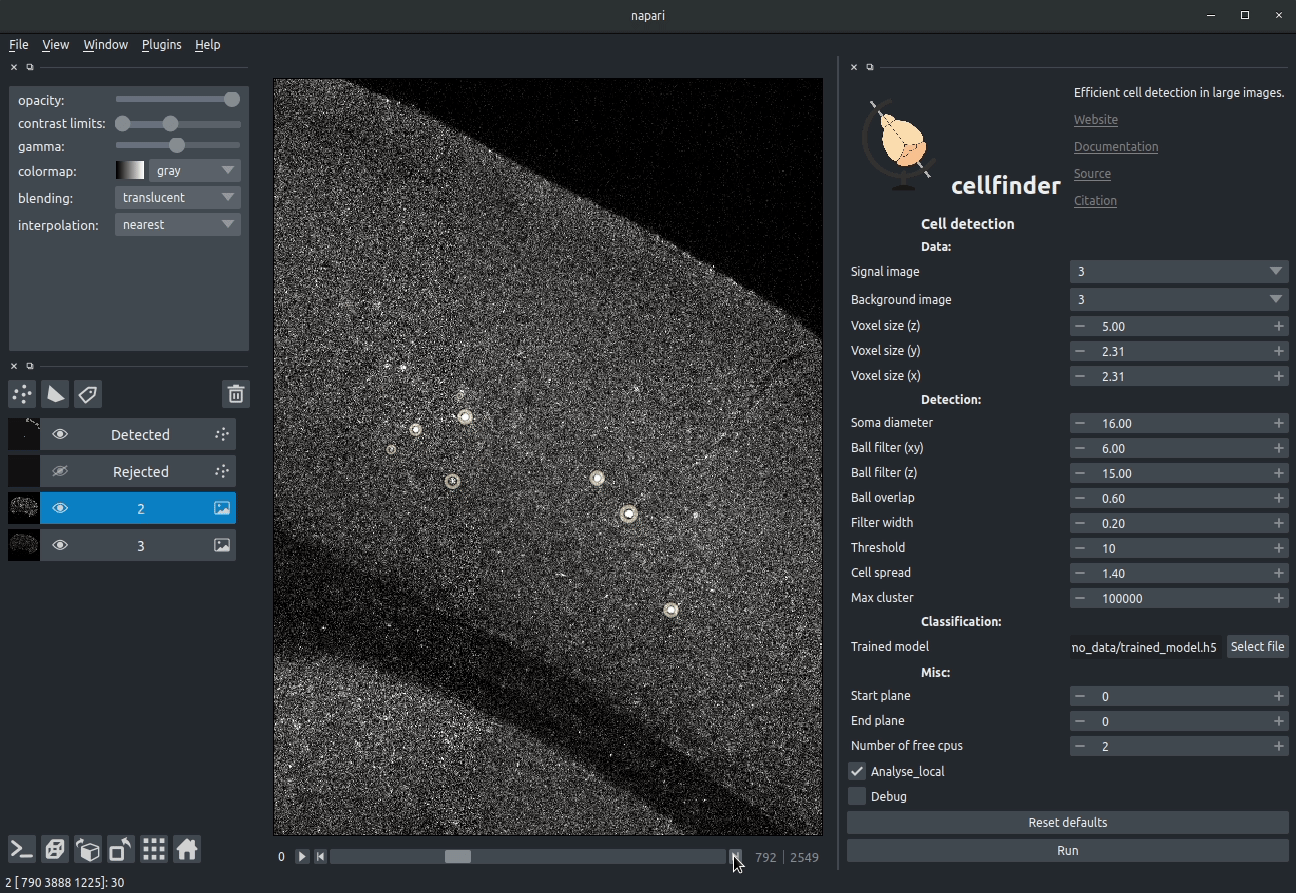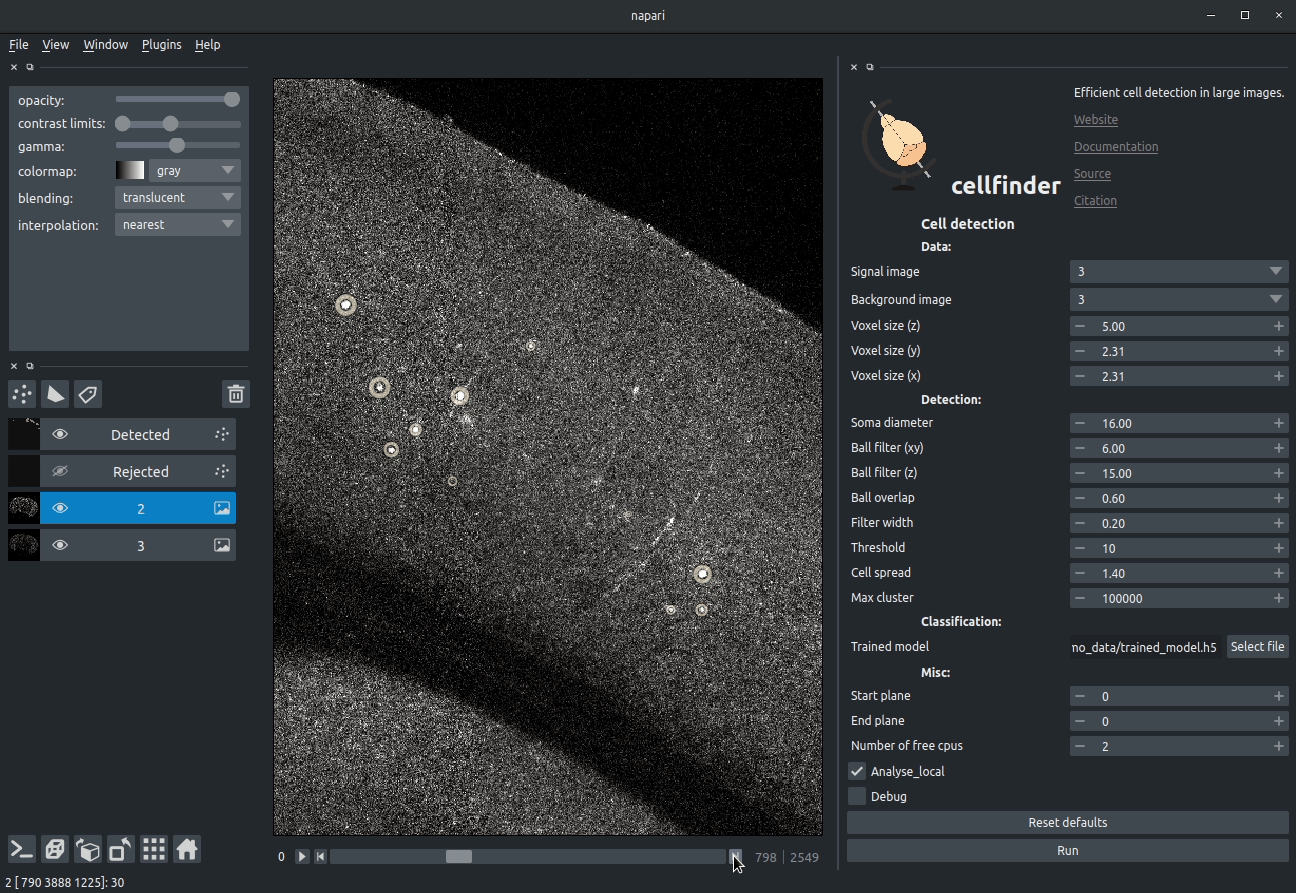
Visualising detected cells in the cellfinder napari plugin
Instructions
Installation
Once you have installed napari.
You can install napari either through the napari plugin installation tool, or
directly from PyPI with:
bash
pip install cellfinder-napari
Usage
Full documentation can be found here.
This software is at a very early stage, and was written with our data in mind. Over time we hope to support other data types/formats. If you have any questions or issues, please get in touch on the forum or by raising an issue.
Illustration
Introduction
cellfinder takes a stitched, but otherwise raw dataset with at least two channels: * Background channel (i.e. autofluorescence) * Signal channel, the one with the cells to be detected:
 Raw coronal serial two-photon mouse brain image showing labelled cells
Raw coronal serial two-photon mouse brain image showing labelled cells
Cell candidate detection
Classical image analysis (e.g. filters, thresholding) is used to find cell-like objects (with false positives):
 Candidate cells (including many artefacts)
Candidate cells (including many artefacts)
Cell candidate classification
A deep-learning network (ResNet) is used to classify cell candidates as true cells or artefacts:
 Cassified cell candidates. Yellow - cells, Blue - artefacts
Cassified cell candidates. Yellow - cells, Blue - artefacts
Contributing
Contributions to cellfinder-napari are more than welcome. Please see the developers guide.
Citing cellfinder
If you find this plugin useful, and use it in your research, please cite the paper outlining the cell detection algorithm:
Tyson, A. L., Rousseau, C. V., Niedworok, C. J., Keshavarzi, S., Tsitoura, C., Cossell, L., Strom, M. and Margrie, T. W. (2021) “A deep learning algorithm for 3D cell detection in whole mouse brain image datasets’ PLOS Computational Biology, 17(5), e1009074 https://doi.org/10.1371/journal.pcbi.1009074
If you use this, or any other tools in the brainglobe suite, please let us know, and we'd be happy to promote your paper/talk etc.
Owner
- Name: BrainGlobe
- Login: brainglobe
- Kind: organization
- Location: London/Munich
- Website: https://brainglobe.info/
- Twitter: brain_globe
- Repositories: 28
- Profile: https://github.com/brainglobe
Open python tools for morphological analyses in systems neuroscience
GitHub Events
Total
Last Year
Committers
Last synced: over 1 year ago
Top Committers
| Name | Commits | |
|---|---|---|
| Adam Tyson | c****e@a****m | 82 |
| alessandrofelder | a****r@u****k | 49 |
| David Stansby | d****y@g****m | 42 |
| paddyroddy | p****y@g****m | 16 |
| pre-commit-ci[bot] | 6****] | 16 |
| Adam Tyson | a****n@u****k | 7 |
| Alessandro Felder | a****r | 3 |
| Will Graham | 3****1 | 3 |
| Justin Kiggins | j****s@g****m | 1 |
Committer Domains (Top 20 + Academic)
Issues and Pull Requests
Last synced: 9 months ago
All Time
- Total issues: 57
- Total pull requests: 74
- Average time to close issues: 3 months
- Average time to close pull requests: 23 days
- Total issue authors: 9
- Total pull request authors: 7
- Average comments per issue: 2.37
- Average comments per pull request: 3.22
- Merged pull requests: 67
- Bot issues: 0
- Bot pull requests: 16
Past Year
- Issues: 0
- Pull requests: 0
- Average time to close issues: N/A
- Average time to close pull requests: N/A
- Issue authors: 0
- Pull request authors: 0
- Average comments per issue: 0
- Average comments per pull request: 0
- Merged pull requests: 0
- Bot issues: 0
- Bot pull requests: 0
Top Authors
Issue Authors
- alessandrofelder (17)
- adamltyson (17)
- dstansby (15)
- chrytsi (2)
- paddyroddy (2)
- viktorpm (1)
- carshadi (1)
- goanpeca (1)
- nal10 (1)
Pull Request Authors
- dstansby (32)
- pre-commit-ci[bot] (16)
- adamltyson (15)
- alessandrofelder (5)
- paddyroddy (3)
- willGraham01 (3)
- MysticElephant (1)
Top Labels
Issue Labels
Pull Request Labels
Packages
- Total packages: 3
-
Total downloads:
- pypi 175 last-month
-
Total dependent packages: 3
(may contain duplicates) -
Total dependent repositories: 1
(may contain duplicates) - Total versions: 48
- Total maintainers: 1
proxy.golang.org: github.com/brainglobe/cellfinder-napari
- Documentation: https://pkg.go.dev/github.com/brainglobe/cellfinder-napari#section-documentation
- License: bsd-3-clause
-
Latest release: v1.0.2
published about 2 years ago
Rankings
pypi.org: cellfinder-napari
Efficient cell detection in large images
- Homepage: https://brainglobe.info/cellfinder
- Documentation: https://docs.brainglobe.info/cellfinder-napari/
- License: BSD-3-Clause
-
Latest release: 1.0.2
published about 2 years ago
Rankings
Maintainers (1)
conda-forge.org: cellfinder-napari
- Homepage: https://brainglobe.info/cellfinder
- License: BSD-3-Clause
-
Latest release: 0.0.20
published over 3 years ago
Rankings
Dependencies
- actions/checkout v2 composite
- chanzuckerberg/napari-hub-preview-action v0.1 composite
- actions/checkout v2 composite
- actions/setup-python v2 composite
- brainglobe/actions/check_manifest v1 composite
- brainglobe/actions/lint v1 composite
- brainglobe/actions/test v1 composite
- tlambert03/setup-qt-libs v1 composite










