https://github.com/brentp/peddy
genotype :: ped correspondence check, ancestry check, sex check. directly, quickly on VCF
Science Score: 13.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
○CITATION.cff file
-
○codemeta.json file
-
○.zenodo.json file
-
✓DOI references
Found 3 DOI reference(s) in README -
○Academic publication links
-
○Committers with academic emails
-
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (13.6%) to scientific vocabulary
Keywords
Keywords from Contributors
Repository
genotype :: ped correspondence check, ancestry check, sex check. directly, quickly on VCF
Basic Info
Statistics
- Stars: 141
- Watchers: 6
- Forks: 40
- Open Issues: 27
- Releases: 0
Topics
Metadata Files
README.md
Fast Pedigree::VCF QC

peddy compares familial-relationships and sexes as reported in a PED/FAM file with those inferred from a VCF.
It samples the VCF at about 25000 sites (plus chrX) to accurately estimate relatedness, IBS0, heterozygosity, sex and ancestry. It uses 2504 thousand genome samples as backgrounds to calibrate the relatedness calculation and to make ancestry predictions.
It does this very quickly by sampling, by using C for computationally intensive parts, and by parallelization.
If you use peddy, please cite Pedersen and Quinlan, Who’s Who? Detecting and Resolving Sample Anomalies in Human DNA Sequencing Studies with Peddy, The American Journal of Human Genetics (2017), http://dx.doi.org/10.1016/j.ajhg.2017.01.017
Note that somalier is a more scalable, faster, replacement for peddy that uses some of the same methods as peddy along with some new ones.
Quickstart
See installation below.
Most users will only need to run as a command-line tool with a ped and VCF, e.g:
python -m peddy -p 4 --plot --prefix ceph-1463 data/ceph1463.peddy.vcf.gz data/ceph1463.ped
This will use 4 cpus to run various checks and create ceph-1463.html which you can open in any browser to interactively explore your data.
It will also create create 4 csv files and 4 QC plots. These will indicate:
- discrepancies between ped-reported and genotype-inferred relations
- discrepancies between ped-reported and genotype-inferred sex
- samples with higher levels of HET calls, lower depth, or more variance in b-allele-frequency (ref / (ref + alt )) for het calls.
- an ancestry prediction based on projection onto the thousand genomes principal components
Finally, it will create a new file ped files ceph1463.peddy.ped that also lists
the most useful columns from the het-check and sex-check. Users can first
look at this extended ped file for an overview of likely problems.
See the docs for a walk-through and thorough explanation of each plot.
hg38 or custom sites
By default, peddy uses hg19/GRCh37. It can be forced to use sites for hg38 by passing --sites hg38.
To create custom sites, have a look at the sites files included with peddy along
with the corresponding .bin.gz which is just the raw binary alternate counts (gt_types) from thousand-genomes that have been written as uint8
and gzipped.
Speed
Because of the sampling approach and parallelization, peddy is very fast.
With 4 CPUs, on the 17-member CEPH1643 pedigree whole-genome VCF, peddy can run the het-check and PCA in ~ 8 seconds. The pedigree check comparing all vs.
all samples run in 3.6 seconds.
It finishes the full set of checks in about 20 seconds.
In comparison KING runs in 14 seconds (it is extremely fast); the time including the conversion from VCF to binary ped is 85 seconds.
On larger datasets, with hundreds or thousands of samples, it can be beneficial to add as many cores as possible; for smaller datasets with dozens of samples about 4 processors reduces the computation time as much as 8 or more would.
Validation
The results between peddy and KING are comparable, but peddy does better on cohorts where most samples are related. See the figure below where the peddy relatedness estimate is closer to the actual than KING which over-estimates relatedness.
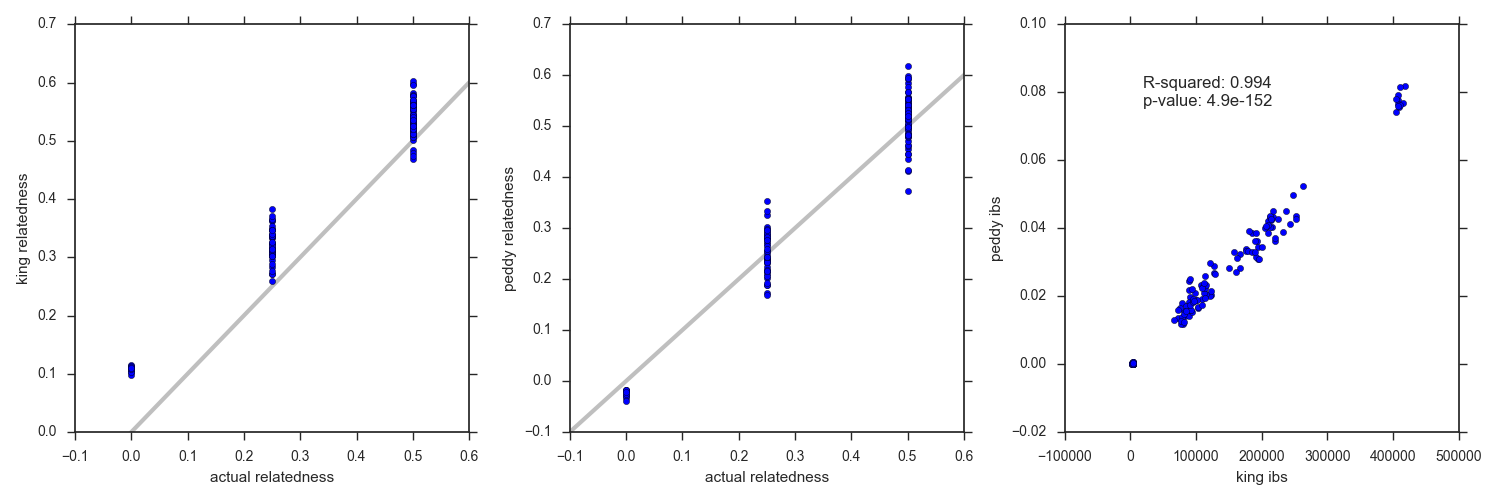
Peddy uses the KING algorithm for calculating relatedness and so they match quite well. Peddy also runs PCA on the 2504 samples from 1000 genomes, then fitting an SVM and predicting ancestry in addition to calculating relatedness among all pairwise combinations of the 17 samples.
Warnings and Checks
On creating a pedigree object (via Ped('some.ped'). peddy will print warnings to STDERR as appropriate like:
``` pedigree warning: '101811-101811' is dad but has female sex pedigree warning: '101897-101897' is dad but has female sex pedigree warning: '101896-101896' is mom of self pedigree warning: '102110-102110' is mom but has male sex pedigree warning: '102110-102110' is mom of self pedigree warning: '101381-101381' is dad but has female sex pedigree warning: '101393-101393' is mom but has male sex
unknown sample: 102498-102498 in family: K34175 unknown sample: 11509-11509 in family: K567331 unknown sample: 5180-5180 in family: K8565 ```
Installation
Conda
Nearly all users should install using conda in the anaconda python distribution. This means have your own version of python easily installed via:
``` INSTALLPATH=~/anaconda wget http://repo.continuum.io/miniconda/Miniconda2-latest-Linux-x8664.sh
or wget http://repo.continuum.io/miniconda/Miniconda2-latest-MacOSX-x86_64.sh
bash Miniconda2-latest* -fbp $INSTALLPATH PATH=$INSTALLPATH/bin:$PATH
conda update -y conda conda config --add channels bioconda
conda install -y peddy ```
This should install all dependencies so you can then run peddy with 4 processes as:
python -m peddy --plot -p 4 --prefix mystudy $VCF $PED
Github
git clone https://github.com/brentp/peddy
cd peddy
pip install -r requirements.txt
pip install --editable .
run with
peddy --plot -p 4 --prefix mystudy $VCF $PED
Owner
- Name: Brent Pedersen
- Login: brentp
- Kind: user
- Location: Oregon, USA
- Twitter: brent_p
- Repositories: 220
- Profile: https://github.com/brentp
Doing genomics
GitHub Events
Total
- Watch event: 6
- Push event: 3
- Fork event: 1
Last Year
- Watch event: 6
- Push event: 3
- Fork event: 1
Committers
Last synced: 9 months ago
Top Committers
| Name | Commits | |
|---|---|---|
| Brent Pedersen | b****e@g****m | 292 |
| Michael Cormier | u****3@k****s | 8 |
| Måns Magnusson | m****n@s****e | 6 |
| gataga | c****s@g****m | 1 |
| Sander Bollen | s****j | 1 |
| Sam Lichtenberg | s****e@g****m | 1 |
| Christian Brueffer | c****n@b****o | 1 |
Committer Domains (Top 20 + Academic)
Issues and Pull Requests
Last synced: 6 months ago
All Time
- Total issues: 81
- Total pull requests: 9
- Average time to close issues: 4 months
- Average time to close pull requests: 12 days
- Total issue authors: 55
- Total pull request authors: 8
- Average comments per issue: 3.25
- Average comments per pull request: 1.78
- Merged pull requests: 8
- Bot issues: 0
- Bot pull requests: 0
Past Year
- Issues: 0
- Pull requests: 0
- Average time to close issues: N/A
- Average time to close pull requests: N/A
- Issue authors: 0
- Pull request authors: 0
- Average comments per issue: 0
- Average comments per pull request: 0
- Merged pull requests: 0
- Bot issues: 0
- Bot pull requests: 0
Top Authors
Issue Authors
- brentp (17)
- henrikstranneheim (3)
- numicator (2)
- snashraf (2)
- liushuangmms (2)
- Fazulur (2)
- angelussong (2)
- arq5x (2)
- claudiadast (2)
- wym0072003 (2)
- leipzig (1)
- N-damo (1)
- UmerMaqsood10 (1)
- bhanratt (1)
- maksjutov (1)
Pull Request Authors
- mikecormier (2)
- moonso (1)
- chapmanb (1)
- cbrueffer (1)
- cassimons (1)
- brentp (1)
- splichte (1)
- sndrtj (1)
Top Labels
Issue Labels
Pull Request Labels
Packages
- Total packages: 1
-
Total downloads:
- pypi 675 last-month
- Total dependent packages: 0
- Total dependent repositories: 7
- Total versions: 27
- Total maintainers: 1
pypi.org: peddy
pleasingly pythonic pedigree manipulation
- Documentation: https://peddy.readthedocs.io/
- License: MIT License
-
Latest release: 0.4.8
published over 4 years ago
Rankings
Maintainers (1)
Dependencies
- sphinx *
- nose * development
- click *
- coloredlogs *
- cython *
- cyvcf2 >=0.5.3
- matplotlib >=1.5.0
- networkx *
- numpy *
- pandas *
- scikit-learn *
- seaborn *
- toolshed *