aidsorb
Python package for deep learning on molecular point clouds.
Science Score: 57.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
✓CITATION.cff file
Found CITATION.cff file -
✓codemeta.json file
Found codemeta.json file -
✓.zenodo.json file
Found .zenodo.json file -
✓DOI references
Found 4 DOI reference(s) in README -
○Academic publication links
-
○Academic email domains
-
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (11.1%) to scientific vocabulary
Keywords
Repository
Python package for deep learning on molecular point clouds.
Basic Info
- Host: GitHub
- Owner: adosar
- License: gpl-3.0
- Language: Python
- Default Branch: master
- Homepage: https://aidsorb.readthedocs.io/en/stable/
- Size: 16.6 MB
Statistics
- Stars: 4
- Watchers: 1
- Forks: 1
- Open Issues: 2
- Releases: 2
Topics
Metadata Files
README.md




 [](https://app.codecov.io/gh/adosar/aidsorb)
[](https://aidsorb.readthedocs.io/en/stable/)
[](https://pypi.org/project/aidsorb/)
[](https://aidsorb-online.streamlit.app)
[](https://app.codecov.io/gh/adosar/aidsorb)
[](https://aidsorb.readthedocs.io/en/stable/)
[](https://pypi.org/project/aidsorb/)
[](https://aidsorb-online.streamlit.app)
AIdsorb is a Python package for deep learning on molecular point clouds.
This package aims to provide a simple, easy-to-use and reproduce interface for:
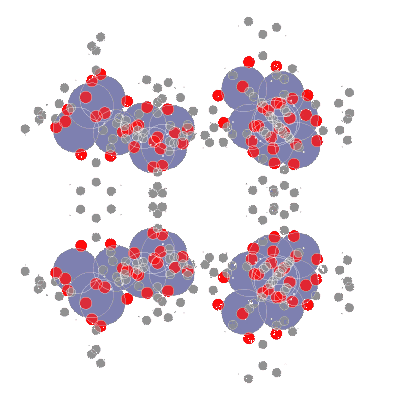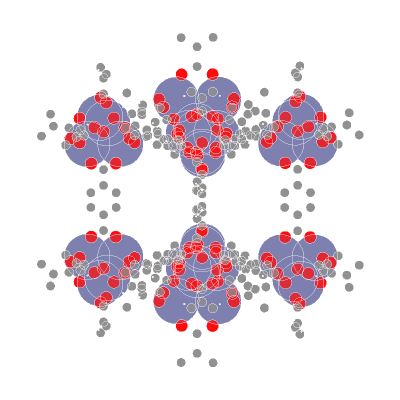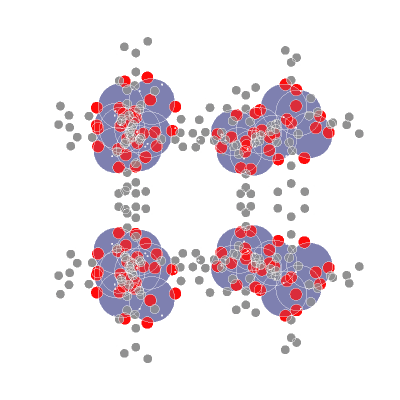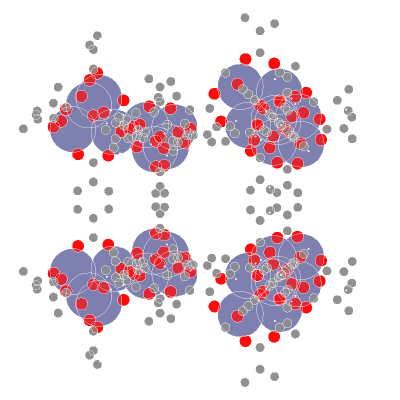
📥 Creating molecular point clouds
🤖 Training DL algorithms on molecular point clouds
⚙️ Installation
[!IMPORTANT] It is strongly recommended to perform the installation inside a virtual environment.
Assuming an activated virtual environment:
bash
pip install aidsorb
🚀 Usage
[!NOTE] Refer to the 📚 Documentation for more information.
Here is a summary of what you can do from the command line:
Visualize a molecular point cloud:
bash aidsorb visualize path/to/structureCreate and prepare point clouds:
bash aidsorb create path/to/structures path/to/pcd_data # Create and store point clouds aidsorb prepare path/to/pcd_data # Split point clouds to train, valdation and testTrain and test a model:
bash aidsorb-lit fit --config=path/to/config.yaml aidsorb-lit test --config=path/to/config.yaml --ckpt_path=path/to/ckpt
💡 Contributing
🙌 We welcome contributions from the community to help improve and expand this project!
You can start by 🛠️ opening an issue for:
- 🐛 Reporting bugs
- 🌟 Suggesting new features
- 📚 Improving documentation
- 🎨 Adding your example to the Gallery
We appreciate your efforts to submit well-documented 🔃 pull requests and participate in discussions.
💪 Together, we can make this project even better!
📑 Citing
- To cite the software, please refer to the citation file or click the citation button.
- To cite the paper, please use the following BibTeX entry:
Show BibTex entry
bibtex
@article{Sarikas2024,
title = {Gas adsorption meets geometric deep learning: points, set and match},
volume = {14},
ISSN = {2045-2322},
url = {http://dx.doi.org/10.1038/s41598-024-76319-8},
DOI = {10.1038/s41598-024-76319-8},
number = {1},
journal = {Scientific Reports},
publisher = {Springer Science and Business Media LLC},
author = {Sarikas, Antonios P. and Gkagkas, Konstantinos and Froudakis, George E.},
year = {2024},
month = nov
}
⚖️ License
AIdosrb is released under the GNU General Public License v3.0 only.
Owner
- Name: Antonios P. Sarikas
- Login: adosar
- Kind: user
- Company: Department of Chemistry, University of Crete
- Repositories: 1
- Profile: https://github.com/adosar
Citation (CITATION.cff)
# This CITATION.cff file was generated with cffinit.
# Visit https://bit.ly/cffinit to generate yours today!
cff-version: 1.2.0
title: AIdsorb
message: >-
If you use this software, please cite it using the
metadata from this file.
type: software
authors:
- given-names: Antonios P.
family-names: Sarikas
orcid: 'https://orcid.org/0009-0008-8420-927X'
identifiers:
- type: doi
value: 10.1038/s41598-024-76319-8
description: The DOI of the paper.
repository-code: 'https://github.com/adosar/aidsorb'
url: 'https://aidsorb.readthedocs.io/en/stable/'
abstract: >-
Python package for deep learning on molecular point
clouds.
keywords:
- deep learning
- geometric deep learning
- point clouds
- machine learning
- pytorch
- pytorch lightning
- porous materials
- material informatics
- metal-organic frameworks
- molecular point clouds
license: GPL-3.0-only
GitHub Events
Total
- Issues event: 112
- Watch event: 4
- Delete event: 2
- Issue comment event: 18
- Push event: 44
- Create event: 2
Last Year
- Issues event: 112
- Watch event: 4
- Delete event: 2
- Issue comment event: 18
- Push event: 44
- Create event: 2
Issues and Pull Requests
Last synced: 6 months ago
All Time
- Total issues: 64
- Total pull requests: 1
- Average time to close issues: 2 days
- Average time to close pull requests: about 1 hour
- Total issue authors: 1
- Total pull request authors: 1
- Average comments per issue: 0.22
- Average comments per pull request: 0.0
- Merged pull requests: 0
- Bot issues: 0
- Bot pull requests: 0
Past Year
- Issues: 63
- Pull requests: 0
- Average time to close issues: 2 days
- Average time to close pull requests: N/A
- Issue authors: 1
- Pull request authors: 0
- Average comments per issue: 0.22
- Average comments per pull request: 0
- Merged pull requests: 0
- Bot issues: 0
- Bot pull requests: 0
Top Authors
Issue Authors
- adosar (64)
Pull Request Authors
- adosar (1)
Top Labels
Issue Labels
Pull Request Labels
Packages
- Total packages: 1
-
Total downloads:
- pypi 166 last-month
- Total dependent packages: 0
- Total dependent repositories: 0
- Total versions: 2
- Total maintainers: 1
pypi.org: aidsorb
Python package for deep learning on molecular point clouds.
- Homepage: https://github.com/adosar/aidsorb
- Documentation: https://aidsorb.readthedocs.io/en/stable/
- License: GPL-3.0-only
-
Latest release: 1.0.0
published over 1 year ago
Rankings
Maintainers (1)
Dependencies
- actions/checkout v4 composite
- actions/setup-python v5 composite
- actions/checkout v4 composite
- actions/download-artifact v4 composite
- actions/setup-python v4 composite
- actions/upload-artifact v4 composite
- pypa/gh-action-pypi-publish release/v1 composite
- sigstore/gh-action-sigstore-python v2.1.1 composite
- actions/checkout v4 composite
- actions/setup-python v5 composite
- ase >=3.22.1
- fire >=0.5.0
- jsonargparse >=4.27.7
- kaleido ==0.2.1
- lightning >=2.2.1
- numpy <=1.26.4
- pandas >=2.2.0
- plotly >=5.19.0
- renku-sphinx-theme ==0.4.0
- scipy >=1.12.0
- sphinx ==7.3.7
- sphinx-copybutton ==0.5.2
- sphinx-design ==0.6.0
- sphinx-gallery ==0.16.0
- tqdm >=4.66.2
- ase >=3.22.1
- fire >=0.5.0
- jsonargparse [signatures]>=4.27.7
- lightning >=2.2.1
- numpy <=1.26.4
- pandas >=2.2.0
- plotly >=5.19.0
- scipy >=1.12.0
- tqdm >=4.66.2