Science Score: 67.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
✓CITATION.cff file
Found CITATION.cff file -
✓codemeta.json file
Found codemeta.json file -
✓.zenodo.json file
Found .zenodo.json file -
✓DOI references
Found 6 DOI reference(s) in README -
○Academic publication links
-
✓Committers with academic emails
1 of 5 committers (20.0%) from academic institutions -
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (13.8%) to scientific vocabulary
Keywords
Keywords from Contributors
Repository
Julia interface to BioMASS
Basic Info
- Host: GitHub
- Owner: biomass-dev
- License: mit
- Language: Julia
- Default Branch: master
- Homepage: https://biomass-dev.github.io/BioMASS.jl/dev
- Size: 1.66 MB
Statistics
- Stars: 0
- Watchers: 1
- Forks: 2
- Open Issues: 0
- Releases: 22
Topics
Metadata Files
README.md
The BioMASS module for Julia
This module provides a Julia interface to the BioMASS parameter estimation.
Installation
The package is a registered package, and can be installed with Pkg.add.
julia
julia> using Pkg; Pkg.add("BioMASS")
or through the pkg REPL mode by typing
] add BioMASS
Python package requirements:
- numpy - https://numpy.org
- scipy - https://scipy.org
- matplotlib - https://matplotlib.org
Example
Model development
This example shows you how to build a simple Michaelis-Menten two-step enzyme catalysis model.
E + S ⇄ ES → E + P
pasmopy.Text2Model allows you to build a BioMASS model from text. You simply describe biochemical reactions and the molecular mechanisms extracted from text are converted into an executable model.
Prepare a text file describing the biochemical reactions (e.g., michaelis_menten.txt)
``` E + S ⇄ ES | kf=0.003, kr=0.001 | E=100, S=50 ES → E + P | kf=0.002
@obs Substrate: u[S] @obs Efree: u[E] @obs Etotal: u[E] + u[ES] @obs Product: u[P] @obs Complex: u[ES]
@sim tspan: [0, 100] ```
Convert the text into an executable model
shell
$ python # pasmopy requires Python 3.7+
```python
from pasmopy import Text2Model description = Text2Model("michaelismenten.txt", lang="julia") description.convert() # generate 'michaelismenten_jl/' ```
Simulate the model using BioMASS.jl
shell
$ julia
```julia using BioMASS
model = Model("./michaelismentenjl"); run_simulation(model) ```
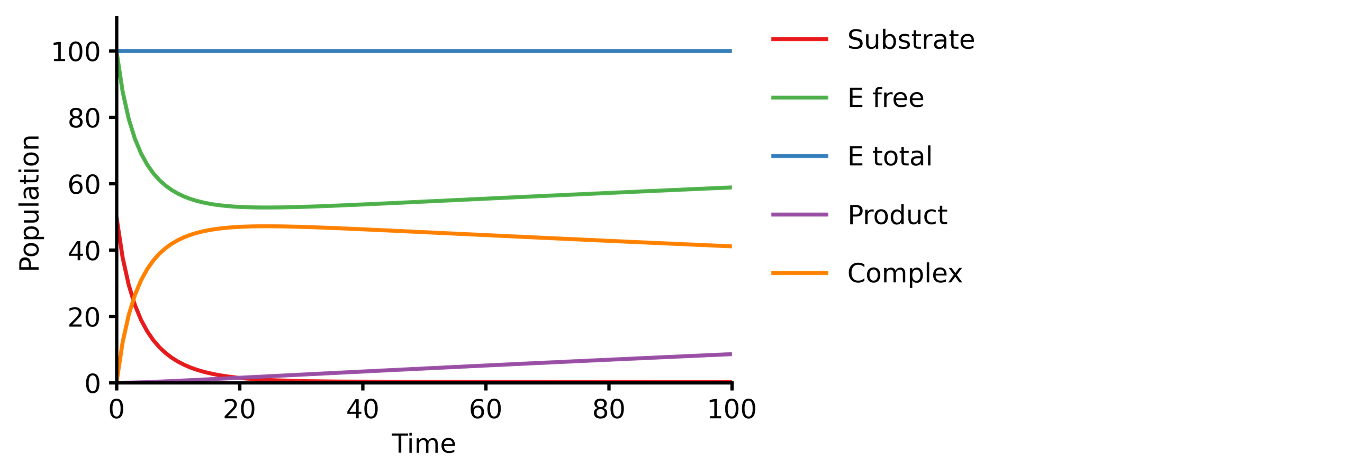
Parameter estimation
```julia using BioMASS
model = Model("./examples/fos_model");
Estimate unknown model parameters from experimental observations
scipydifferentialevolution(model, 1) # requires scipy package
Save simulation results to figure/ in the model folder
runsimulation(model, viztype="best", show_all=true) ```
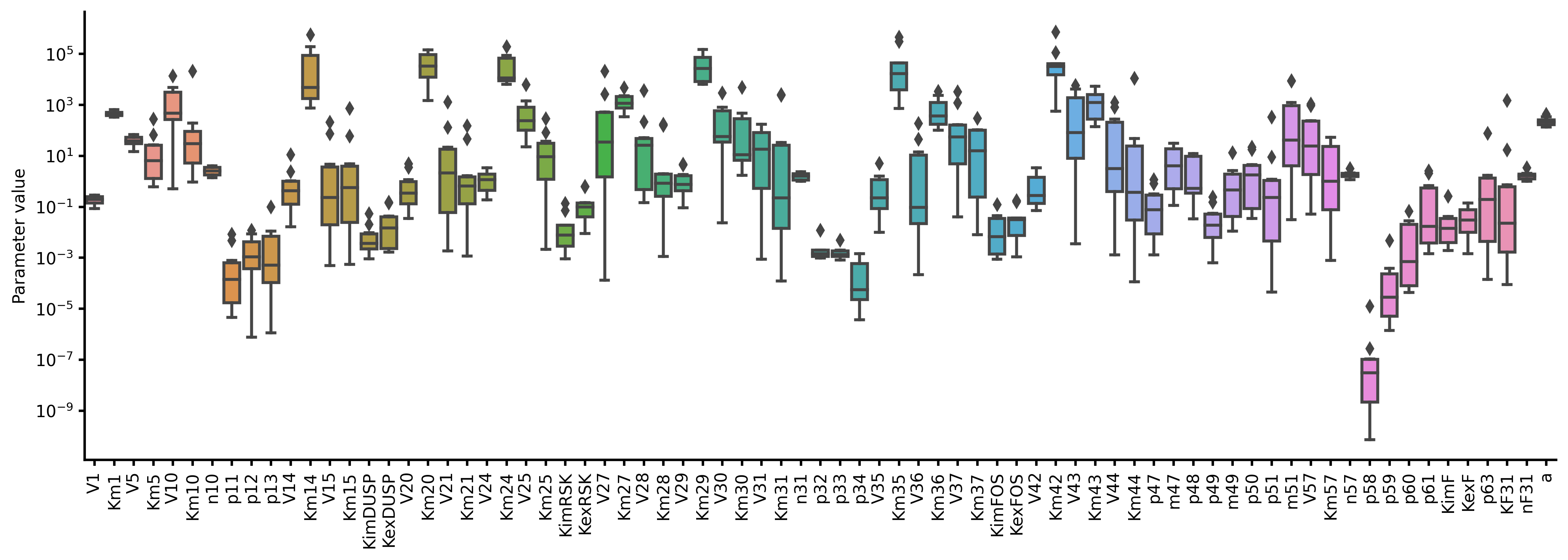
References
Imoto, H., Zhang, S. & Okada, M. A Computational Framework for Prediction and Analysis of Cancer Signaling Dynamics from RNA Sequencing Data—Application to the ErbB Receptor Signaling Pathway. Cancers 12, 2878 (2020). https://doi.org/10.3390/cancers12102878
Imoto, H., Yamashiro, S. & Okada, M. A text-based computational framework for patient -specific modeling for classification of cancers. iScience 25, 103944 (2022). https://doi.org/10.1016/j.isci.2022.103944
License
Owner
- Name: BioMASS
- Login: biomass-dev
- Kind: organization
- Repositories: 2
- Profile: https://github.com/biomass-dev
Open-source software project providing tools for modeling and analysis of biological signaling systems.
Citation (CITATION.cff)
cff-version: 1.2.0
message: "If you use this software in your work, please cite the following paper:"
preferred-citation:
type: article
authors:
- family-names: "Imoto"
given-names: "Hiroaki"
orcid: "https://orcid.org/0000-0002-6817-642X"
- family-names: "Zhang"
given-names: "Suxiang"
- family-names: "Okada"
given-names: "Mariko"
orcid: "https://orcid.org/0000-0002-6210-8223"
doi: "10.3390/cancers12102878"
journal: "Cancers"
title: "A Computational Framework for Prediction and Analysis of Cancer Signaling Dynamics from RNA Sequencing Data—Application to the ErbB Receptor Signaling Pathway"
issue: 10
volume: 12
start: 2878
end: 2878
year: 2020
url: "https://www.mdpi.com/2072-6694/12/10/2878"
GitHub Events
Total
Last Year
Committers
Last synced: almost 3 years ago
All Time
- Total Commits: 213
- Total Committers: 5
- Avg Commits per committer: 42.6
- Development Distribution Score (DDS): 0.15
Top Committers
| Name | Commits | |
|---|---|---|
| himoto | 3****o@u****m | 181 |
| github-actions[bot] | 4****]@u****m | 14 |
| Hiroaki Imoto | h****o@p****p | 14 |
| CompatHelper Julia | c****y@j****g | 2 |
| Hiroaki Imoto | h****o@u****e | 2 |
Committer Domains (Top 20 + Academic)
Issues and Pull Requests
Last synced: 6 months ago
All Time
- Total issues: 5
- Total pull requests: 63
- Average time to close issues: 6 days
- Average time to close pull requests: 5 days
- Total issue authors: 2
- Total pull request authors: 2
- Average comments per issue: 8.0
- Average comments per pull request: 0.0
- Merged pull requests: 60
- Bot issues: 0
- Bot pull requests: 20
Past Year
- Issues: 0
- Pull requests: 0
- Average time to close issues: N/A
- Average time to close pull requests: N/A
- Issue authors: 0
- Pull request authors: 0
- Average comments per issue: 0
- Average comments per pull request: 0
- Merged pull requests: 0
- Bot issues: 0
- Bot pull requests: 0
Top Authors
Issue Authors
- himoto (4)
- JuliaTagBot (1)
Pull Request Authors
- himoto (43)
- github-actions[bot] (19)
Top Labels
Issue Labels
Pull Request Labels
Packages
- Total packages: 1
- Total downloads: unknown
- Total dependent packages: 0
- Total dependent repositories: 0
- Total versions: 14
juliahub.com: BioMASS
Julia interface to BioMASS
- Homepage: https://biomass-dev.github.io/BioMASS.jl/dev
- Documentation: https://docs.juliahub.com/General/BioMASS/stable/
- License: MIT
-
Latest release: 0.8.2
published almost 3 years ago
Rankings
Dependencies
- actions/checkout v2 composite
- julia-actions/setup-julia latest composite
- JuliaRegistries/TagBot v1 composite
- actions/checkout v2 composite
- julia-actions/julia-buildpkg v1 composite
- julia-actions/julia-runtest v1 composite
- julia-actions/setup-julia v1 composite



