gmr
gmr: Gaussian Mixture Regression - Published in JOSS (2021)
Science Score: 100.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
✓CITATION.cff file
Found CITATION.cff file -
✓codemeta.json file
Found codemeta.json file -
✓.zenodo.json file
Found .zenodo.json file -
✓DOI references
Found 15 DOI reference(s) in README and JOSS metadata -
✓Academic publication links
Links to: joss.theoj.org, zenodo.org -
✓Committers with academic emails
1 of 4 committers (25.0%) from academic institutions -
○Institutional organization owner
-
✓JOSS paper metadata
Published in Journal of Open Source Software
Keywords
Scientific Fields
Repository
Gaussian Mixture Regression
Basic Info
- Host: GitHub
- Owner: AlexanderFabisch
- License: bsd-3-clause
- Language: Python
- Default Branch: master
- Homepage: https://alexanderfabisch.github.io/gmr/
- Size: 2.35 MB
Statistics
- Stars: 197
- Watchers: 5
- Forks: 52
- Open Issues: 4
- Releases: 8
Topics
Metadata Files
README.md
gmr
Gaussian Mixture Models (GMMs) for clustering and regression in Python.
- Source code repository: https://github.com/AlexanderFabisch/gmr
- License: New BSD / BSD 3-clause
- Releases: https://github.com/AlexanderFabisch/gmr/releases
- API documentation
Documentation
Installation
Install from PyPI:
bash
pip install gmr
If you want to be able to run all examples, pip can install all necessary examples with
bash
pip install gmr[all]
You can also install gmr from source:
bash
pip install -e .
Example
Estimate GMM from samples, sample from GMM, and make predictions:
```python import numpy as np from gmr import GMM
Your dataset as a NumPy array of shape (nsamples, nfeatures):
X = np.random.randn(100, 2)
gmm = GMM(ncomponents=3, randomstate=0) gmm.from_samples(X)
Estimate GMM with expectation maximization:
X_sampled = gmm.sample(100)
Make predictions with known values for the first feature:
x1 = np.random.randn(20, 1) x1index = [0] x2predictedmean = gmm.predict(x1index, x1) ```
For more details, see:
python
help(gmr)
or have a look at the API documentation
You can see the results of all the examples here.
You can find worked examples in this Google Colab notebook.
How Does It Compare to scikit-learn?
There is an implementation of Gaussian Mixture Models for clustering in scikit-learn as well. Regression could not be easily integrated in the interface of sklearn. That is the reason why I put the code in a separate repository. It is possible to initialize GMR from sklearn though:
python
from sklearn.mixture import GaussianMixture
from gmr import GMM
gmm_sklearn = GaussianMixture(n_components=3, covariance_type="diag")
gmm_sklearn.fit(X)
gmm = GMM(
n_components=3, priors=gmm_sklearn.weights_, means=gmm_sklearn.means_,
covariances=np.array([np.diag(c) for c in gmm_sklearn.covariances_]))
For model selection with sklearn we furthermore provide an optional regressor interface.
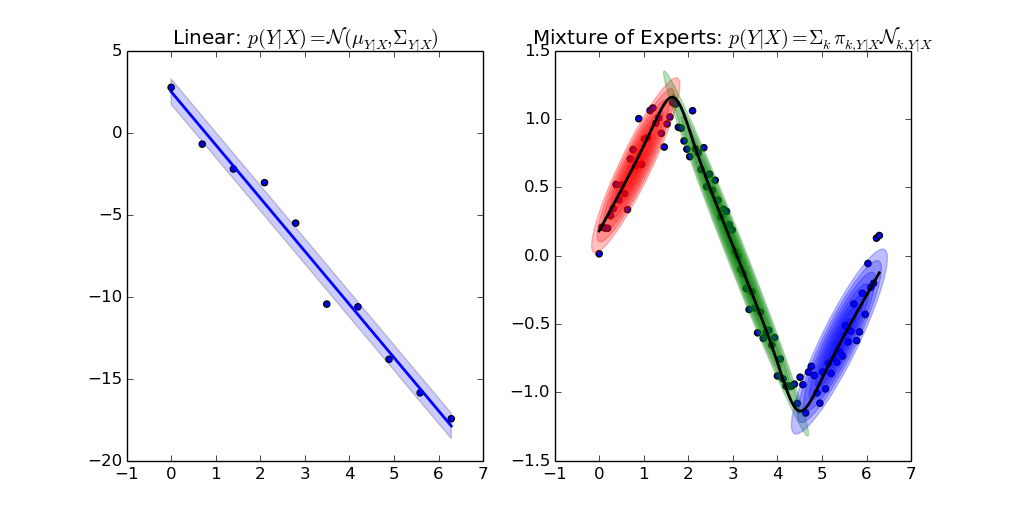
Gallery
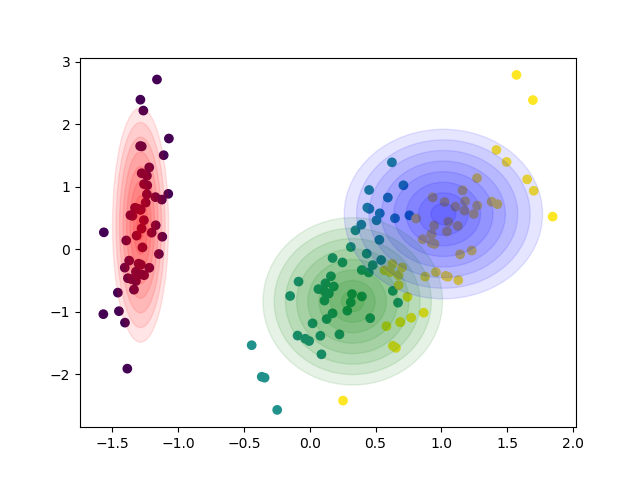
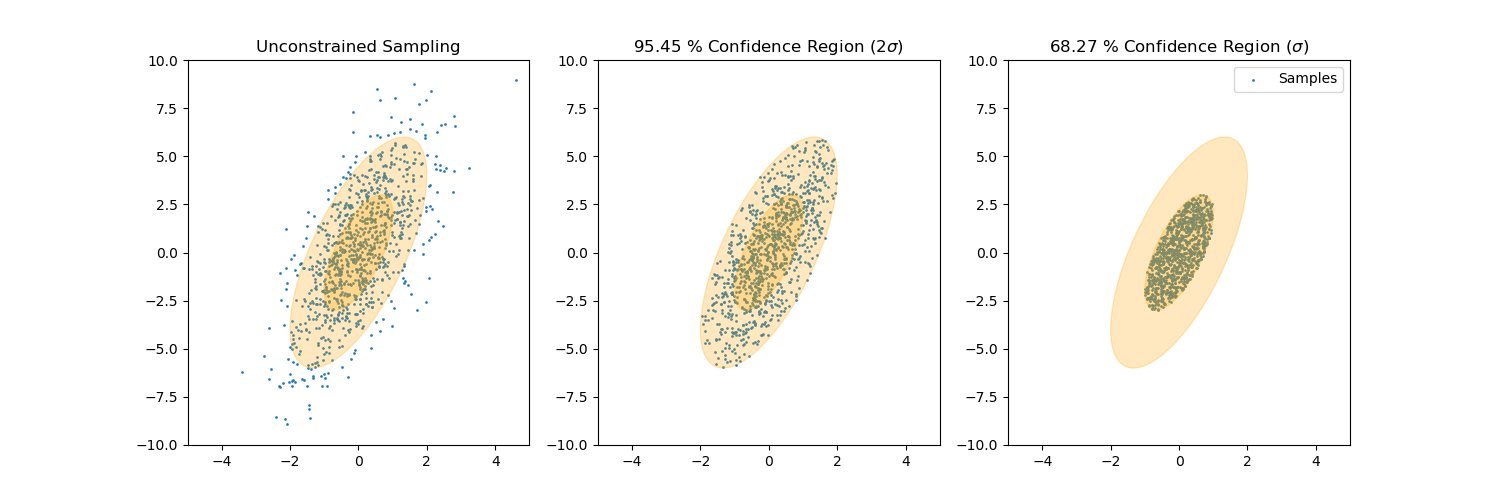
Sample from confidence interval
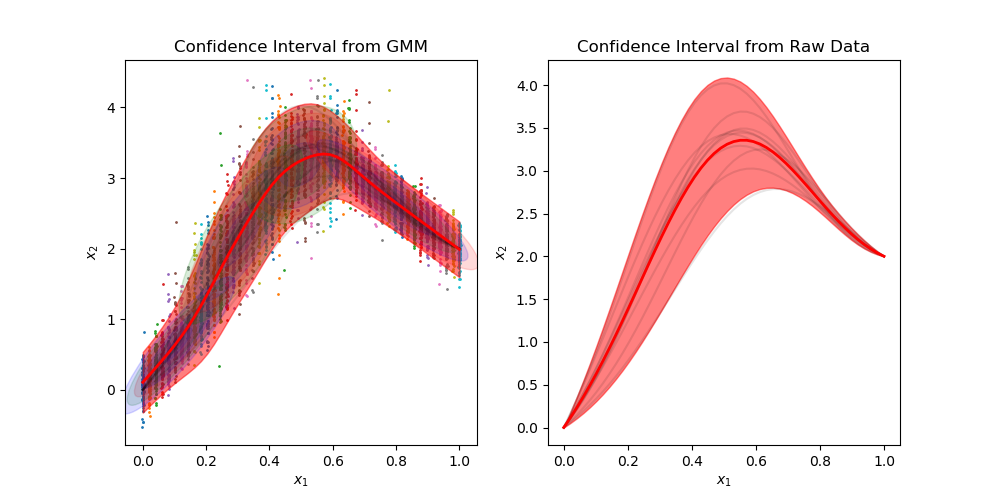
Sample time-invariant trajectories
You can find all examples here.
Saving a Model
This library does not directly offer a function to store fitted models. Since the implementation is pure Python, it is possible, however, to use standard Python tools to store Python objects. For example, you can use pickle to temporarily store a GMM:
```python import numpy as np import pickle import gmr gmm = gmr.GMM(ncomponents=2) gmm.fromsamples(X=np.random.randn(1000, 3))
Save object gmm to file 'file'
pickle.dump(gmm, open("file", "wb"))
Load object from file 'file'
gmm2 = pickle.load(open("file", "rb")) ```
It might be required to store models more permanently than in a pickle file,
which might break with a change of the library or with the Python version.
In this case you can choose a storage format that you like and store the
attributes gmm.priors, gmm.means, and gmm.covariances. These can be
used in the constructor of the GMM class to recreate the object and they can
also be used in other libraries that provide a GMM implementation. The
MVN class only needs the attributes mean and covariance to define the
model.
API Documentation
API documentation is available here.
Citation
If you use the library gmr in a scientific publication, I would appreciate citation of the following paper:
Fabisch, A., (2021). gmr: Gaussian Mixture Regression. Journal of Open Source Software, 6(62), 3054, https://doi.org/10.21105/joss.03054
Bibtex entry:
bibtex
@article{Fabisch2021,
doi = {10.21105/joss.03054},
url = {https://doi.org/10.21105/joss.03054},
year = {2021},
publisher = {The Open Journal},
volume = {6},
number = {62},
pages = {3054},
author = {Alexander Fabisch},
title = {gmr: Gaussian Mixture Regression},
journal = {Journal of Open Source Software}
}
Contributing
How can I contribute?
If you discover bugs, have feature requests, or want to improve the documentation, you can open an issue at the issue tracker of the project.
If you want to contribute code, please open a pull request via
GitHub by forking the project, committing changes to your fork,
and then opening a
pull request
from your forked branch to the main branch of gmr.
Development Environment
I would recommend to install gmr from source in editable mode with pip and
install all dependencies:
bash
pip install -e .[all,test,doc]
You can now run tests with
bash
pytest
This will also generate a coverage report and output an HTML overview to
the folder htmlcov/.
Generate Documentation
The API documentation is generated with pdoc3. If you want to regenerate it, you can run
bash
pdoc gmr --html --skip-errors
Related Publications
The first publication that presents the GMR algorithm is
[1] Z. Ghahramani, M. I. Jordan, "Supervised learning from incomplete data via an EM approach," Advances in Neural Information Processing Systems 6, 1994, pp. 120-127, https://proceedings.neurips.cc/paper/1993/hash/f2201f5191c4e92cc5af043eebfd0946-Abstract.html
but it does not use the term Gaussian Mixture Regression, which to my knowledge occurs first in
[2] S. Calinon, F. Guenter and A. Billard, "On Learning, Representing, and Generalizing a Task in a Humanoid Robot," in IEEE Transactions on Systems, Man, and Cybernetics, Part B (Cybernetics), vol. 37, no. 2, 2007, pp. 286-298, doi: 10.1109/TSMCB.2006.886952.
A recent survey on various regression models including GMR is the following:
[3] F. Stulp, O. Sigaud, "Many regression algorithms, one unified model: A review," in Neural Networks, vol. 69, 2015, pp. 60-79, doi: 10.1016/j.neunet.2015.05.005.
Sylvain Calinon has a good introduction in his slides on nonlinear regression for his machine learning course.
Owner
- Name: Alexander Fabisch
- Login: AlexanderFabisch
- Kind: user
- Location: Bremen
- Company: German Research Center for Artificial Intelligence (DFKI GmbH, @dfki), Robotics Innovation Center (@dfki-ric)
- Website: https://alexanderfabisch.github.io/
- Repositories: 62
- Profile: https://github.com/AlexanderFabisch
researcher, software developer, in robotics and machine learning
JOSS Publication
gmr: Gaussian Mixture Regression
Tags
machine learning regressionCitation (CITATION.cff)
cff-version: 1.2.0
message: "If you use this software, please cite it as below."
authors:
- family-names: Fabisch
given-names: Alexander
orcid: https://orcid.org/0000-0003-2824-7956
title: "gmr"
url: https://github.com/AlexanderFabisch/gmr
doi: 10.5281/zenodo.4449631
preferred-citation:
type: article
authors:
- family-names: Fabisch
given-names: Alexander
orcid: https://orcid.org/0000-0003-2824-7956
title: "gmr: Gaussian Mixture Regression"
doi: 10.21105/joss.03054
journal: Journal of Open Source Software
start: 3054
issue: 62
volume: 6
month: 6
year: 2021
Papers & Mentions
Total mentions: 1
Drosophila melanogaster as a Model Organism of Brain Diseases
- DOI: 10.3390/ijms10020407
- OpenAlex ID: https://openalex.org/W2035847800
- Published: February 2009
GitHub Events
Total
- Create event: 2
- Release event: 2
- Issues event: 2
- Watch event: 25
- Issue comment event: 13
- Push event: 13
- Pull request review event: 3
- Pull request review comment event: 1
- Pull request event: 3
- Fork event: 3
Last Year
- Create event: 2
- Release event: 2
- Issues event: 2
- Watch event: 25
- Issue comment event: 13
- Push event: 13
- Pull request review event: 3
- Pull request review comment event: 1
- Pull request event: 3
- Fork event: 3
Committers
Last synced: 7 months ago
Top Committers
| Name | Commits | |
|---|---|---|
| Alexander Fabisch | a****h@g****m | 248 |
| John Kitchin | j****n@a****u | 5 |
| Marcelo R. Albuquerque | m****e@g****m | 4 |
| Joao Felipe Santos | j****l@g****m | 2 |
Committer Domains (Top 20 + Academic)
Issues and Pull Requests
Last synced: 6 months ago
All Time
- Total issues: 31
- Total pull requests: 21
- Average time to close issues: about 2 months
- Average time to close pull requests: 18 days
- Total issue authors: 21
- Total pull request authors: 4
- Average comments per issue: 3.48
- Average comments per pull request: 1.71
- Merged pull requests: 18
- Bot issues: 0
- Bot pull requests: 0
Past Year
- Issues: 0
- Pull requests: 3
- Average time to close issues: N/A
- Average time to close pull requests: about 12 hours
- Issue authors: 0
- Pull request authors: 2
- Average comments per issue: 0
- Average comments per pull request: 3.33
- Merged pull requests: 2
- Bot issues: 0
- Bot pull requests: 0
Top Authors
Issue Authors
- AlexanderFabisch (10)
- nardi (2)
- mralbu (1)
- MetaDev (1)
- inakleinbottle (1)
- nikolai-neustroev (1)
- ostwalprasad (1)
- wout-konings (1)
- rocreguant (1)
- dmronga (1)
- Labulitiolle (1)
- xlzhu0317 (1)
- show0k (1)
- meshiguge (1)
- sdodingnan (1)
Pull Request Authors
- AlexanderFabisch (17)
- jkitchin (4)
- mralbu (1)
- jfsantos (1)
- ARoefer (1)
Top Labels
Issue Labels
Pull Request Labels
Packages
- Total packages: 1
-
Total downloads:
- pypi 103,218 last-month
- Total dependent packages: 6
- Total dependent repositories: 5
- Total versions: 11
- Total maintainers: 3
pypi.org: gmr
Gaussian Mixture Regression
- Homepage: https://github.com/AlexanderFabisch/gmr
- Documentation: https://gmr.readthedocs.io/
- License: bsd-3-clause
-
Latest release: 2.0.2
published 6 months ago
Rankings
Maintainers (3)
Dependencies
- numpy *
- actions/checkout v2 composite
- actions/setup-python v2 composite
- codecov/codecov-action v1.3.2 composite
