ivadomed
ivadomed: A Medical Imaging Deep Learning Toolbox - Published in JOSS (2021)
Science Score: 95.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
○CITATION.cff file
-
✓codemeta.json file
Found codemeta.json file -
✓.zenodo.json file
Found .zenodo.json file -
✓DOI references
Found 4 DOI reference(s) in README and JOSS metadata -
✓Academic publication links
Links to: joss.theoj.org -
✓Committers with academic emails
1 of 35 committers (2.9%) from academic institutions -
○Institutional organization owner
-
✓JOSS paper metadata
Published in Journal of Open Source Software
Keywords from Contributors
Scientific Fields
Repository
Repository on the collaborative IVADO medical imaging project between the Mila and NeuroPoly labs.
Basic Info
- Host: GitHub
- Owner: ivadomed
- License: mit
- Language: Python
- Default Branch: master
- Homepage: https://ivadomed.org
- Size: 134 MB
Statistics
- Stars: 163
- Watchers: 10
- Forks: 147
- Open Issues: 302
- Releases: 34
Metadata Files
README.md
[!WARNING]
ivadomedis no more maintained. New models integrated in our 3rd party software (SCT, AxonDeepSeg, etc.) are now trained using MONAI and/or nnUnet.
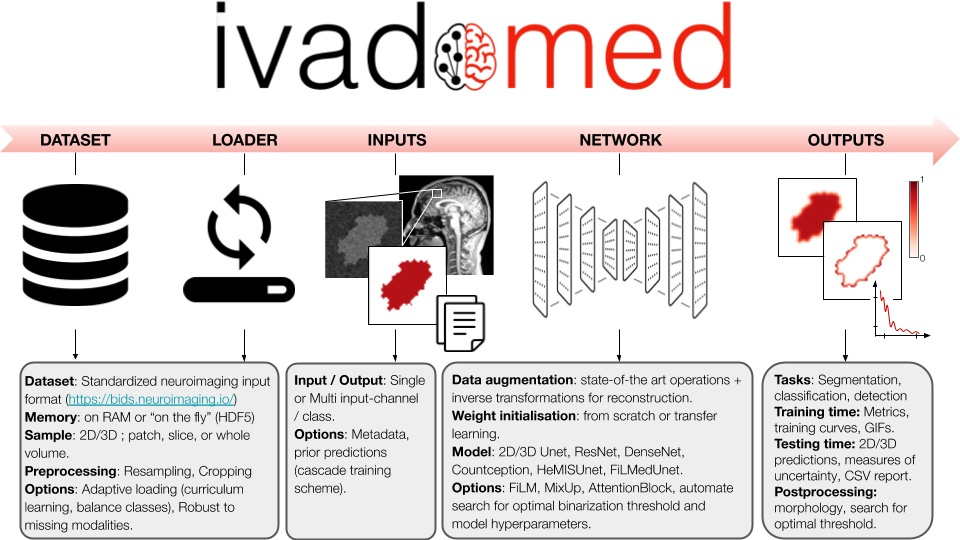
ivadomed is an integrated framework for medical image analysis with deep learning.
The technical documentation is available here. The more detailed installation instruction is available there
Installation
ivadomed requires Python >= 3.7 and < 3.10 as well as PyTorch == 1.8. We recommend working under a virtual environment, which could be set as follows:
bash
python -m venv ivadomed_env
source ivadomed_env/bin/activate
Install from release (recommended)
Install ivadomed and its requirements from Pypi <https://pypi.org/project/ivadomed/>__:
bash
pip install --upgrade pip
pip install ivadomed
Install from source
Bleeding-edge developments builds are available on the project's master branch on Github. Installation procedure is the following:
bash
git clone https://github.com/neuropoly/ivadomed.git
cd ivadomed
pip install -e .
Contributors



This project results from a collaboration between the NeuroPoly Lab and the Mila. The main funding source is IVADO.
Consult our Wiki(https://github.com/ivadomed/ivadomed/wiki) here for more help
Owner
- Name: ivadomed
- Login: ivadomed
- Kind: organization
- Location: Canada
- Website: https://ivadomed.org
- Twitter: ivadomed
- Repositories: 23
- Profile: https://github.com/ivadomed
Integrated framework for medical image analysis with deep learning.
JOSS Publication
ivadomed: A Medical Imaging Deep Learning Toolbox
Authors

NeuroPoly Lab, Institute of Biomedical Engineering, Polytechnique Montreal, Montreal, Canada, Mila, Quebec AI Institute, Montreal, QC, Canada

NeuroPoly Lab, Institute of Biomedical Engineering, Polytechnique Montreal, Montreal, Canada, Mila, Quebec AI Institute, Montreal, QC, Canada

NeuroPoly Lab, Institute of Biomedical Engineering, Polytechnique Montreal, Montreal, Canada, Mila, Quebec AI Institute, Montreal, QC, Canada
NeuroPoly Lab, Institute of Biomedical Engineering, Polytechnique Montreal, Montreal, Canada

NeuroPoly Lab, Institute of Biomedical Engineering, Polytechnique Montreal, Montreal, Canada
NeuroPoly Lab, Institute of Biomedical Engineering, Polytechnique Montreal, Montreal, Canada
Mila, Quebec AI Institute, Montreal, QC, Canada, AIMI, Stanford University, Stanford, CA, USA
Tags
Deep Learning Medical Imaging Segmentation Open-source PipelineGitHub Events
Total
- Issues event: 5
- Watch event: 8
- Issue comment event: 65
- Push event: 6
- Pull request review comment event: 1
- Pull request event: 5
- Gollum event: 1
Last Year
- Issues event: 5
- Watch event: 8
- Issue comment event: 65
- Push event: 6
- Pull request review comment event: 1
- Pull request event: 5
- Gollum event: 1
Committers
Last synced: 7 months ago
Top Committers
| Name | Commits | |
|---|---|---|
| charleygros | c****s@g****m | 1,752 |
| andreanne-lemay | a****y@p****a | 676 |
| Julien Cohen-Adad | j****n@p****a | 239 |
| lrouhier | l****r@t****r | 223 |
| olivier | o****h@h****m | 210 |
| AnBucquet | b****t@e****r | 104 |
| cakester | h****a@m****m | 67 |
| mariehbourget | 5****t | 57 |
| ValentineLL | v****l@e****a | 36 |
| Yang Ding | y****g@m****m | 31 |
| Alexandru Jora | a****u@j****a | 26 |
| Ainsleigh Hill | a****l@g****m | 25 |
| Nick | n****r@p****a | 21 |
| Christian S. Perone | c****e@g****m | 17 |
| Giselle Martel | 3****l | 12 |
| Yuan Wang | 3****n | 12 |
| BUCQUET Anthime | 3****t | 11 |
| Joshua Newton | j****n@g****m | 11 |
| Kanishk Kalra | 3****6 | 9 |
| Uzay Macar | u****r@g****m | 8 |
| Srishti | s****7@g****m | 6 |
| Olivier Vincent | o****n@p****a | 6 |
| Mathieu Boudreau, PhD | e****0@g****m | 5 |
| Konstantinos | k****s@h****m | 4 |
| Naga Karthik | 5****k | 4 |
| bsauty | 4****y | 3 |
| Alexandru Foias | a****s | 3 |
| Enoch Tetteh | c****h@g****m | 1 |
| Fernando França | f****f@a****r | 1 |
| Jeff | n****o@g****m | 1 |
| and 5 more... | ||
Committer Domains (Top 20 + Academic)
Issues and Pull Requests
Last synced: 6 months ago
All Time
- Total issues: 63
- Total pull requests: 174
- Average time to close issues: about 1 month
- Average time to close pull requests: 2 months
- Total issue authors: 13
- Total pull request authors: 26
- Average comments per issue: 3.33
- Average comments per pull request: 3.4
- Merged pull requests: 145
- Bot issues: 0
- Bot pull requests: 0
Past Year
- Issues: 2
- Pull requests: 1
- Average time to close issues: about 9 hours
- Average time to close pull requests: N/A
- Issue authors: 2
- Pull request authors: 1
- Average comments per issue: 0.5
- Average comments per pull request: 2.0
- Merged pull requests: 0
- Bot issues: 0
- Bot pull requests: 0
Top Authors
Issue Authors
- jcohenadad (41)
- naga-karthik (4)
- joshuacwnewton (4)
- mathieuboudreau (3)
- mariehbourget (2)
- uzaymacar (2)
- hermancollin (2)
- kiristern (2)
- taimast (1)
- dbsi-pinkman (1)
- zwb0 (1)
- chennyyuee (1)
Pull Request Authors
- andreanne-lemay (34)
- mariehbourget (18)
- ahill187 (16)
- jcohenadad (15)
- cakester (15)
- joshuacwnewton (14)
- charleygros (13)
- kanishk16 (8)
- mathieuboudreau (8)
- lrouhier (6)
- kaidalisohaib (5)
- gisellemartel (5)
- kousu (4)
- mpompolas (3)
- konantian (2)
Top Labels
Issue Labels
Pull Request Labels
Dependencies
- csv-diff >=1.0
- joblib *
- loguru *
- matplotlib >=3.3.0
- nibabel *
- onnxruntime ==1.7.0
- pandas *
- pillow >=7.0.0
- pybids >=0.14.0
- pyparsing <3,>=2.0.2
- scikit-image *
- scikit-learn >=0.20.3
- scipy *
- seaborn *
- tensorboard >=1.15.0
- torch *
- torchio >=0.18.68
- torchvision *
- tqdm >=4.30
- wandb >=0.12.11
- actions/cache v3 composite
- actions/checkout v3 composite
- actions/setup-python v3 composite
- actions/checkout v3 composite
- actions/setup-python v3 composite
- pypa/gh-action-pypi-publish release/v1 composite
- softprops/action-gh-release v1 composite
- actions/checkout v3 composite
- actions/setup-python v3 composite
- pip
- python >=3.7,<3.9

