Science Score: 36.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
○CITATION.cff file
-
✓codemeta.json file
Found codemeta.json file -
✓.zenodo.json file
Found .zenodo.json file -
○DOI references
-
○Academic publication links
-
✓Committers with academic emails
1 of 2 committers (50.0%) from academic institutions -
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (15.4%) to scientific vocabulary
Keywords
Repository
RepeatFS
Basic Info
Statistics
- Stars: 8
- Watchers: 1
- Forks: 1
- Open Issues: 3
- Releases: 0
Topics
Metadata Files
README.md
RepeatFS
RepeatFS: a file system providing reproducibility through provenance and automation
RepeatFS is a Python, FUSE-based file system with the goal of promoting scientific informatics reproducibility by recording all file and IO operations during analysis. This provenance record can then be exported and used to replicate the analysis on other systems. During replication, RepeatFS will verify that all software versions, command line parameters, and other relevant attributes match, and will report any deviation from the original record. While other provenance software often involves learning scripting languages or migrating your workflow to a confined platform, RepeatFS operates invisibly at the file system level and is compatible with virtually all Linux command-line software.
In addition to replication and verification, RepeatFS also provides provenance visualization. It is capable of generating a webpage visualizing the complete provenance history, including all programs that wrote to that file, all files read by those programs, all programs that wrote to those programs, etc.
Lastly, RepeatFS provides Virtual Dynamic Files (VDFs). These VDFs automatically execute commonly performed tasks (such as converting file types) in a systematic and uniform fashion. Each supported file will have a corresponding VDF presented on disk. Upon accessing this file, RepeatFS will run the appropriate task, and populate the file on the fly with the correct information. These files are then cached in memory, so subsequently accessing them does not require the process to run a second time. VDFs may be chained together for combining operations and may be copied to turn them into normal files.
INSTALLATION
Dependencies * Python 3 (and pip) * FUSE 2 (and libfuse) * GraphViz
Dependencies are available in all popular system package managers. Python 3, Libfuse, and GraphViz are also available within Anaconda (you may be able to find a channel with FUSE available as well):
- Debian/Ubuntu:
sudo apt install python3 python3-pip fuse2 libfuse2 graphviz libgraphviz-dev - RHEL/Fedora:
sudo yum install python3 python3-pip fuse2 fuse2-libs graphviz graphviz-devel - Anaconda:
conda install -c conda-forge python=3 pip libfuse=2 graphviz
After the dependencies have been installed, RepeatFS can be installed from PyPI using pip:
pip3 install repeatfs
If installing using a normal user account, pip will likely install RepeatFS into your ~/.local/bin directory. If this directory is in your PATH variable, you can simply run repeatfs. If not, you'll need to add this directory to your PATH variable, or run RepeatFS using the full path ~/.local/bin/repeatfs.
USAGE
RepeatFS functions as a transparent layer between you and your files, recording all IO activity. In order to use RepeatFS, you'll mount the target directory you want to monitor (which includes subdirectories and files). Then, anytime you wish to access any files within the monitored directory, you'll instead use the path to the RepeatFS mount.
Mount and monitor a directory:
repeatfs mount <directory to monitor> <RepeatFS mount directory>
Stop monitoring a directory:
fusermount -u <RepeatFS mount directory>
The most powerful feature of RepeatFS is the ability to record provenance and replicate the creation of the file on a different system. To ensure all operations are successfully recorded, be sure to perform the entirety of your analysis using a RepeatFS mount.
Path to a file's provenance record - this is a VDF, and is populated automatically when accessed, and may be copied to any location. Note the plus sign next to the file name below - all VDFs are available using a plus sign next to the filename:
<RepeatFS mount directory>/<any sub directories>/<file name>+/<file name>.provenance.json
Replicate a file (replication destination must be within an active RepeatFS mount:
repeatfs replicate -r <replication destination> <provenance file>
Path to a file's provenance graph - like the provenance record, this is also a VDF. RepeatFS visualizes provenance by generating an HTML file that can be viewed in any browser:
<RepeatFS mount directory>/<any sub directories>/<file name>+/<file name>.provenance.html
REPLICATION EXAMPLES
In this example, we first mount our work directory using RepeatFS. Then we download a copy of the UniProt SwissProt database, decompress it, and perform two simple tasks: extract fasta header sequences, and count the number of lines in the fasta file. We take both results, and add it to a new archive.
``` repeatfs mount ~/work ~/mnt cd ~/mnt
wget ftp://ftp.uniprot.org/pub/databases/uniprot/currentrelease/knowledgebase/complete/uniprotsprot.fasta.gz gzip -d uniprotsprot.fasta.gz grep ">" uniprotsprot.fasta > ~/mnt/headers.txt cat uniprot_sprot.fasta | wc -l > ~/mnt/count.txt tar -cvf results.tar headers.txt count.txt ```
To visualize the provenance of the results.tar file within our browser:
cd results.tar+
firefox results.tar.provenance.html
`
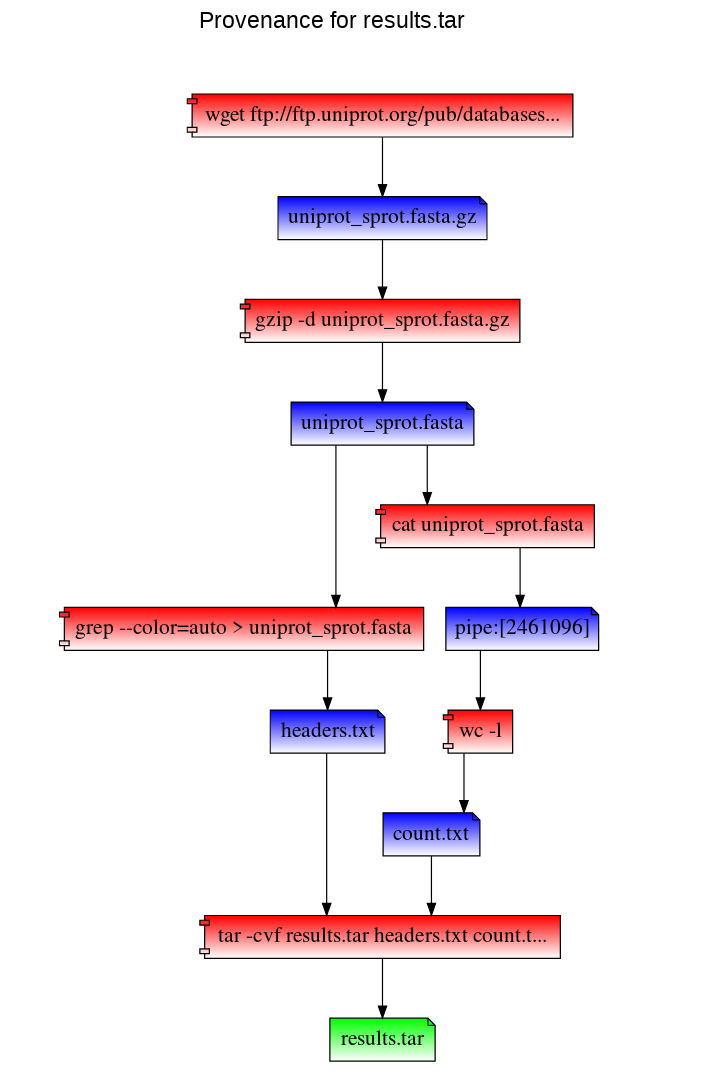
RepeatFS can also replicate these steps to recreate results.tar using the results.tar.provenance.json file. You can use this file (or distribute it to others) to reproduce your work. In the following example, we've copied the provenance record into our home directory. We then mount a directory with RepeatFS and replicate the work (and save stdout and stderr into log files):
``` repeatfs mount ~/replicate ~/mnt cd ~/mnt
repeatfs replicate ~/results.tar.provenance.json --stdout stdout.log --stderr stderr.log ```
RepeatFS will execute and verify each step. Version mismatches or other errors will be reported:
[info] Starting replication
[info] Replication complete
[info] Starting verification
[ok] Process 16056 (wget ftp://ftp.uniprot.org/pub/databases/uniprot/current_release/knowledgebase/complete/uniprot_sprot.fasta.gz) executed
[ok] Process 16091 (gzip -d uniprot_sprot.fasta.gz) executed
[ok] Process 16107 (grep --color=auto > uniprot_sprot.fasta) executed
[ok] Process 16110 (cat uniprot_sprot.fasta) executed
[ok] Process 16113 (wc -l) executed
[ok] Process 16118 (tar -cvf results.tar headers.txt count.txt) executed
[info] Verification complete
RepeatFS can also simply list the commands that would be used during replication (using the -l argument):
repeatfs replicate ~/results.tar.provenance.json -l
This will list each command that will be run, in order. It will also list ID(s) next to each command, which can be used during replication (using the -e argument) to reconstruct a missing shell script.
[turing|1591668563.97|1652] wget ftp://ftp.uniprot.org/pub/databases/uniprot/current_release/knowledgebase/complete/uniprot_sprot.fasta.gz
[turing|1591668614.24|1695] gzip -d uniprot_sprot.fasta.gz
[turing|1591668694.23|1756] grep --color=auto > uniprot_sprot.fasta > /tmp/mnt/headers.txt
[turing|1591669111.77|2165, turing|1591669111.77|2166] cat uniprot_sprot.fasta | wc -l > /tmp/mnt/count.txt
[turing|1591669123.5|2175] tar -cvf results.tar headers.txt count.txt
VIRTUAL DYNAMIC FILES
Provenance and replication are only a part of RepeatFS's capabilities. RepeatFS can also automate commonly performed tasks using VDFs. VDFs are special files representing the output of some operation, such as converting file formats, extracting text, indexing a reference, etc. Whenever RepeatFS detects a file that is a valid input for one of these types of operations, it will also show a corresponding output file. When this output file is accessed (opened, copied, read), RepeatFS will automatically run the program necessary to perform the action, and populate the output file in realtime. These VDFs look and act just like normal files, though they are stored in memory. VDFs may be converted into normal files simply by copying them to another directory.
VDFs are configured within the RepeatFS configuration file as follows:
```
Comments start with
The following entry creates a VDF that shows a corresponding FASTA for any FASTQ.
Accessing the FASTA will automatically run seqtk
[entry] # Each VDF entry starts with [entry] match=.fastq$ # This regular expression controls which files are valid input ext=.fasta # This extension will be appended to the end of the VDF cmd=seqtk seq -A {input} # This is the command that will be run when accessing the VDF ```
All VDF files, including system-provided ones like provenance, or user-defined ones like the FASTQ->FASTA example above, can be found by placing a plus sign + after the input filename. So if we had a FASTQ file located at ~/mnt/example.fastq, listing the contents of ~/mnt/example.fastq+ would show the following files:
example.fastq.fasta
example.fastq.provenance.html
example.fastq.provenance.json
Note that since VDFs are treated as normal files, they also can be used as inputs to other VDFs. In this way, multiple VDFs can be chained together to perform automated operations in a modular fashion. Using the above example, if we define the following VDF:
[entry]
match=.*
ext=.count
cmd=wc -l {input}
This would create a corresponding .count for every file in the RepeatFS mount, containing the number of lines the original file contained (even if that original file was a VDF). In this way, we can view the number of lines in a FASTA converted from the FASTQ file above:
less ~/mnt/example.fastq+/example.fastq.fasta+/example.fastq.fasta.count
While VDFs are not required to record provenance in RepeatFS, they do represent a powerful tool to aid in improving reproducibility by performing common tasks in a uniform, documented fashion.
PLUGINS
RepeatFS supports expanding its functionality through the use of plugins. Plugins are enabled by including the plugins option in the configuration file and specifying the names, comma delimited, of the plugins you wish to enable:
plugins=snapshot
Currently, the snapshot plugin is the only publicly available plugin, although others are planned. The snapshot plugin will capture the contents of specific files and file types when those files are accessed. The file contents are saved into the provenance database and can be viewed/downloaded using the provenance web interface noted above. After selecting the file node in the provenance graph, the info window on the right will display links for the dates and times of each available snapshot. This is especially useful when multiple modifications were made to an analysis script, and you want to verify what particular version of a script was used to generate a result file.
To configure which files you'd like to snapshot, include the snapshot.select option in the configuration file, specifying a regular expression of all the file names/types you'd like to capture.
snapshot.select=\.(py|sh|txt|r|m)$
[!CAUTION] Be careful not to select file types that may be very large, as this may significantly slow down processing while capturing the snapshot and will cause the database size to grow quickly.
If you wish to develop your own plugin, see the template.txt in the plugins subdirectory for further instructions.
QUESTIONS
RepeatFS is actively being developed and improved all the time. If you encounter any issue at all, please don't hesitate to reach out and file a bug report here: https://github.com/ToniWestbrook/repeatfs/issues
Owner
- Name: Toni Westbrook
- Login: ToniWestbrook
- Kind: user
- Location: Portsmouth, NH
- Company: University of New Hampshire
- Repositories: 2
- Profile: https://github.com/ToniWestbrook
Deputy Directory of Research Computing at the University of New Hampshire. Providing bioinformatics, HPC, and general computing solutions.
GitHub Events
Total
- Issue comment event: 2
- Push event: 4
- Pull request event: 3
Last Year
- Issue comment event: 2
- Push event: 4
- Pull request event: 3
Committers
Last synced: over 2 years ago
Top Committers
| Name | Commits | |
|---|---|---|
| Toni Westbrook | a****k@u****u | 59 |
| Toni Westbrook | m****z@j****t | 6 |
Committer Domains (Top 20 + Academic)
Issues and Pull Requests
Last synced: 6 months ago
All Time
- Total issues: 3
- Total pull requests: 4
- Average time to close issues: 22 minutes
- Average time to close pull requests: 2 months
- Total issue authors: 1
- Total pull request authors: 1
- Average comments per issue: 0.67
- Average comments per pull request: 0.5
- Merged pull requests: 1
- Bot issues: 0
- Bot pull requests: 0
Past Year
- Issues: 0
- Pull requests: 4
- Average time to close issues: N/A
- Average time to close pull requests: 2 months
- Issue authors: 0
- Pull request authors: 1
- Average comments per issue: 0
- Average comments per pull request: 0.5
- Merged pull requests: 1
- Bot issues: 0
- Bot pull requests: 0
Top Authors
Issue Authors
- ToniWestbrook (3)
Pull Request Authors
- noahcui (4)
Top Labels
Issue Labels
Pull Request Labels
Packages
- Total packages: 1
-
Total downloads:
- pypi 14 last-month
- Total dependent packages: 0
- Total dependent repositories: 1
- Total versions: 2
- Total maintainers: 1
pypi.org: repeatfs
File system providing reproducibility through provenance and automation
- Homepage: http://github.com/ToniWestbrook/repeatfs
- Documentation: https://repeatfs.readthedocs.io/
- License: LICENSE
-
Latest release: 0.10.0
published over 5 years ago
Rankings
Maintainers (1)
Dependencies
- fusepy *
- pygraphviz *
- python-daemon *