NCBImeta
NCBImeta: efficient and comprehensive metadata retrieval from NCBI databases - Published in JOSS (2020)
Science Score: 93.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
○CITATION.cff file
-
✓codemeta.json file
Found codemeta.json file -
✓.zenodo.json file
Found .zenodo.json file -
✓DOI references
Found 3 DOI reference(s) in README and JOSS metadata -
✓Academic publication links
Links to: joss.theoj.org -
○Committers with academic emails
-
○Institutional organization owner
-
✓JOSS paper metadata
Published in Journal of Open Source Software
Scientific Fields
Repository
Efficient and comprehensive metadata acquisition from the NCBI databases (includes SRA).
Basic Info
- Host: GitHub
- Owner: ktmeaton
- License: mit
- Language: Python
- Default Branch: master
- Homepage: https://ktmeaton.github.io/NCBImeta/
- Size: 26.2 MB
Statistics
- Stars: 50
- Watchers: 3
- Forks: 8
- Open Issues: 3
- Releases: 21
Metadata Files
README.md
Efficient and comprehensive metadata acquisition from NCBI databases (includes SRA).
Why NCBImeta?
NCBImeta is a command-line application that retrieves and organizes metadata from the National Centre for Biotechnology Information (NCBI). While the NCBI web browser experience allows filtered searches, the output does not facilitate inter-record comparison or bulk record retrieval. NCBImeta tackles this issue by creating a local database of NCBI metadata constructed by user-defined search criteria and customizable metadata columns. The output of NCBImeta, optionally a SQLite database or text files, can then be used by computational biologists for applications such as record filtering, project discovery, sample interpretation, or meta-analyses of published work.
Requirements
- NCBImeta is written in Python 3 and supported on Linux and macOS.
- Dependencies that will be installed are listed in requirements.txt.
- Python Versions:
- 3.7
- 3.8
- 3.9
- Operating Systems:
- Ubuntu
- macOS
Conda is the recommended installation method. To install with pip, gcc is required.
Installation
There are three installation options for NCBImeta:
1. Conda*
*
mambais strongly recommended overconda!
bash
conda env create -f environment.yaml
conda activate ncbimeta
2. PyPI*
*
gccis required.
bash
pip install ncbimeta
3. Github
bash
git clone https://github.com/ktmeaton/NCBImeta.git
cd NCBImeta
pip install .
Test that the installation was successful:
bash
NCBImeta --version
Command-Line Parameters
```text usage: NCBImeta [-h] --config CONFIGPATH [--flat] [--version] [--email USEREMAIL] [--api USERAPI] [--force-pause-seconds USERFORCEPAUSESECONDS]
NCBImeta: Efficient and comprehensive metadata retrieval from the NCBI databases.
optional arguments:
-h, --help show this help message and exit
--config CONFIGPATH Path to the yaml configuration file (ex. config.yaml).
--flat Don't create sub-directories in output directory.
--version show program's version number and exit
--email USEREMAIL User email to override parameter in config file.
--api USERAPI User API key to override parameter in config file.
--force-pause-seconds USERFORCEPAUSESECONDS
FORCE PAUSE SECONDS to override parameter in config
file.
--quiet Suppress logging of each record to the console.
```
Quick Start Example
Access the quick start config file
Download the NCBImeta github repository to get access to the example configuration files:
bash
git clone https://github.com/ktmeaton/NCBImeta.git
cd NCBImeta
Run the program
Download a selection of genomic metadata pertaining to the plague pathogen Yersinia pestis.
bash
NCBImeta --flat --config test/test.yaml
(Note: The 'quick' start config file forces slow downloads to accommodate users with slow internet. For faster record retrieval, please see the config file docs to start editing config files.)
Example output of the command-line interface (v0.6.1):
Annotate the database with the user's custom metadata
bash
NCBImetaAnnotate \
--database test/test.sqlite \
--annotfile test/test_annot.txt \
--table BioSample
Note that the first column of your annotation file MUST be a column that is unique to each record. An Accession number or ID is highly recommended. The column headers in your annotation file must also exactly match the names of your columns in the database.
NCBImetaAnnotate by default replaces the existing annotation with the data in your custom metadata file. Alternatively, the flag --concatenate can be specified. This will concatenate your custom metadata with the pre-existing value in the database cell (separated by a semi-colon).
bash
NCBImetaAnnotate \
--database test/test.sqlite \
--annotfile test/test_annot.txt \
--table BioSample \
--concatenate
Join NCBI tables into a unified master table
bash
NCBImetaJoin \
--database test/test.sqlite \
--final Master \
--anchor BioSample \
--accessory "BioProject Assembly SRA Nucleotide" \
--unique "BioSampleAccession BioSampleAccessionSecondary BioSampleBioProjectAccession"
The rows of the output "Master" table will be from the anchor table "BioSample", with additional columns added in from the accessory tables "BioProject", "Assembly", "SRA", and "Nucleotide". Unique accession numbers for BioSample (both primary and secondary) and BioProject allow this join to be unambiguous.
Export the database to tab-separated text files by table.
bash
NCBImetaExport \
--database test/test.sqlite \
--outputdir test
Each table within the database will be exported to its own tab-separated .txt file in the specified output directory.
Explore!
- Explore your database text files using a spreadsheet viewer (Microsoft Excel, Google Sheets, etc.)
- Browse your SQLite database using DB Browser for SQLite (https://sqlitebrowser.org/)
- Use the columns with FTP links to download your data files of interest.
Example database output (a subset of the BioSample table)
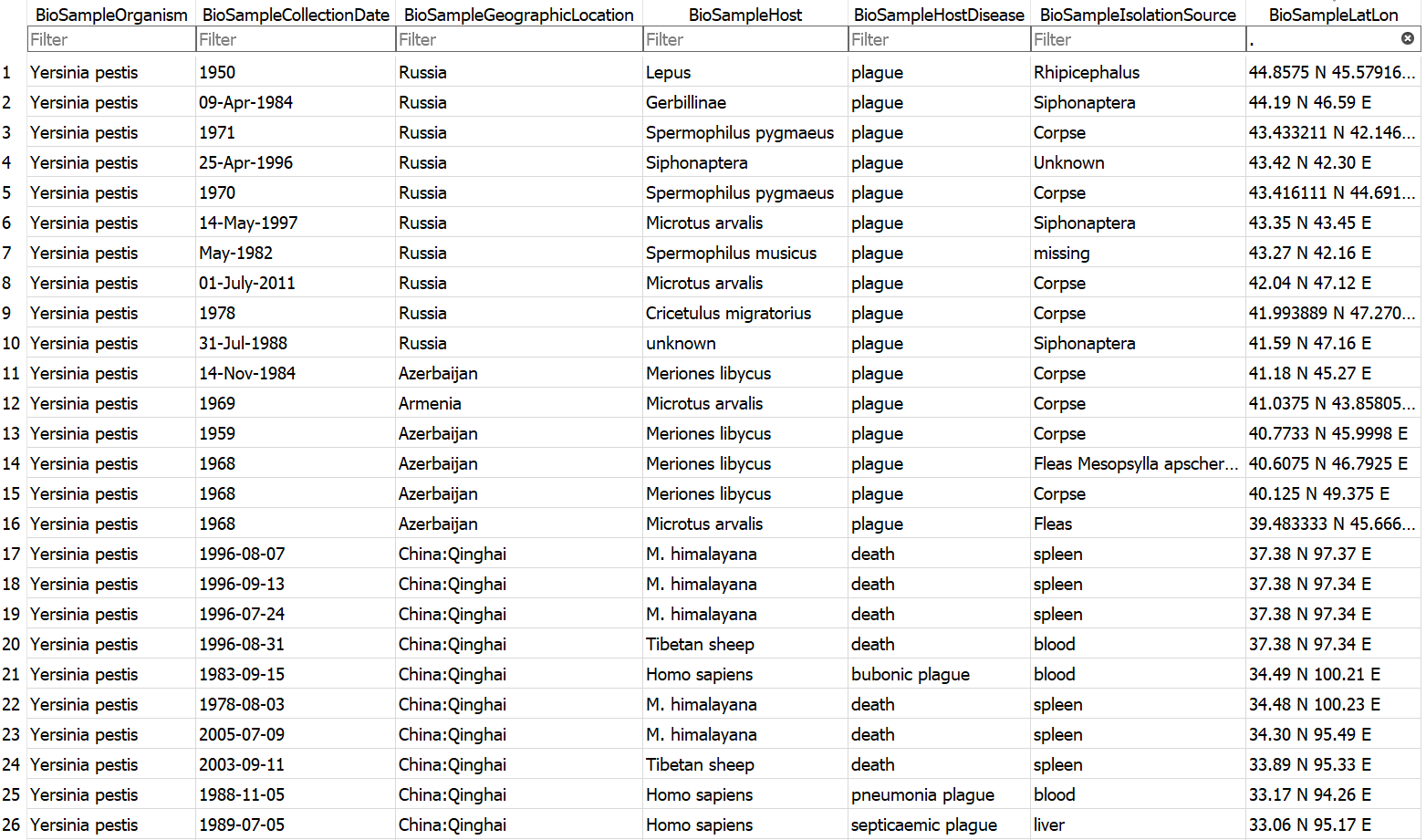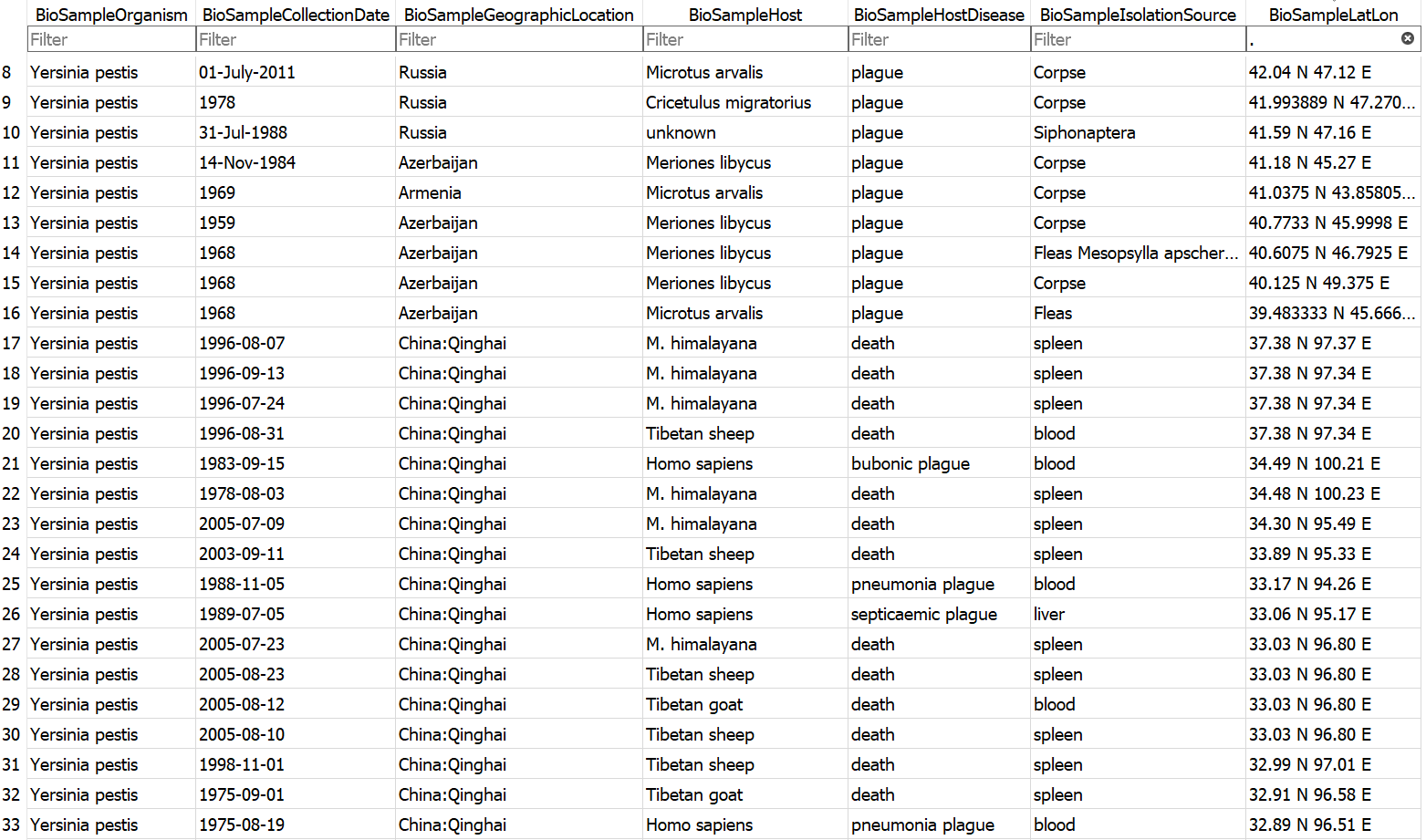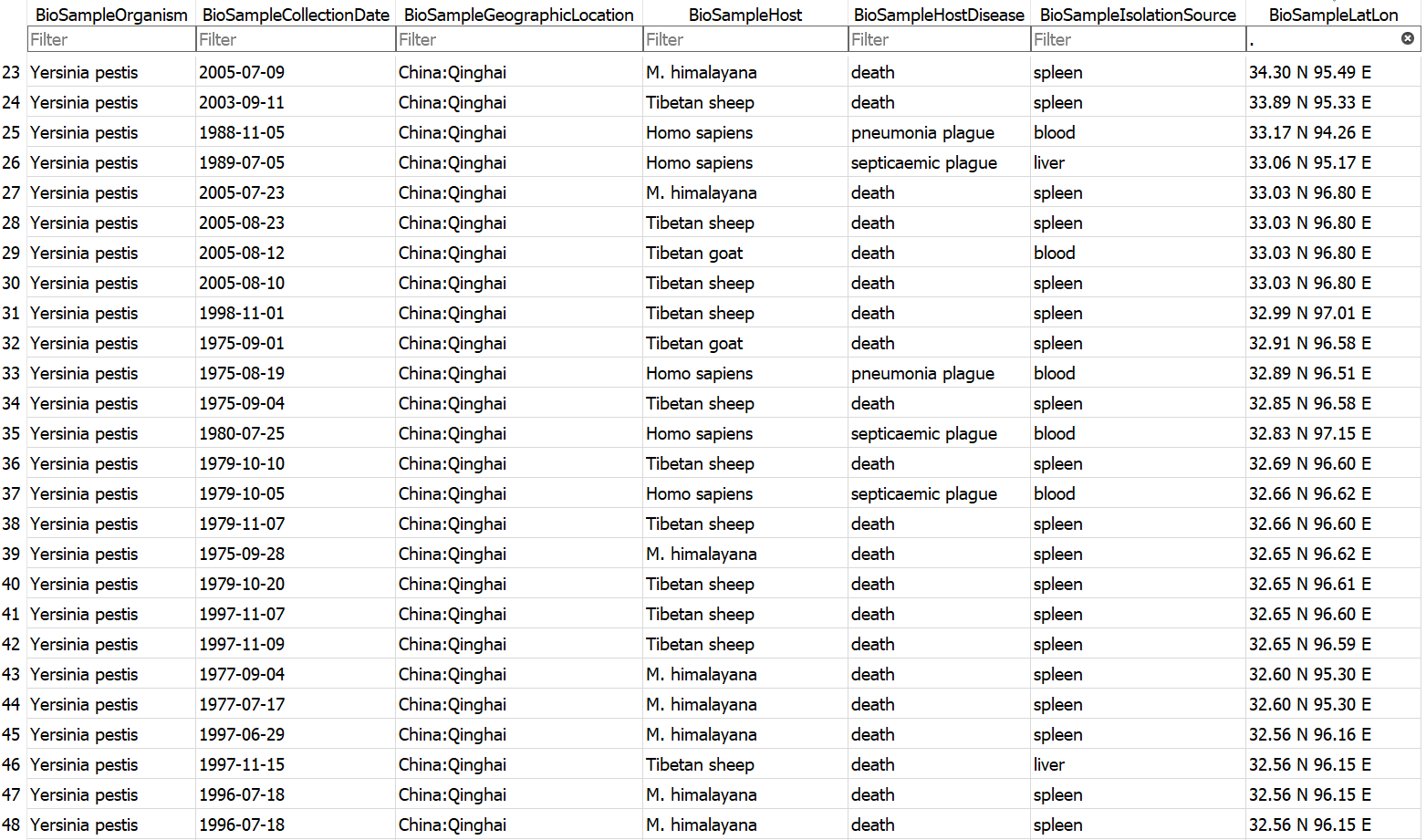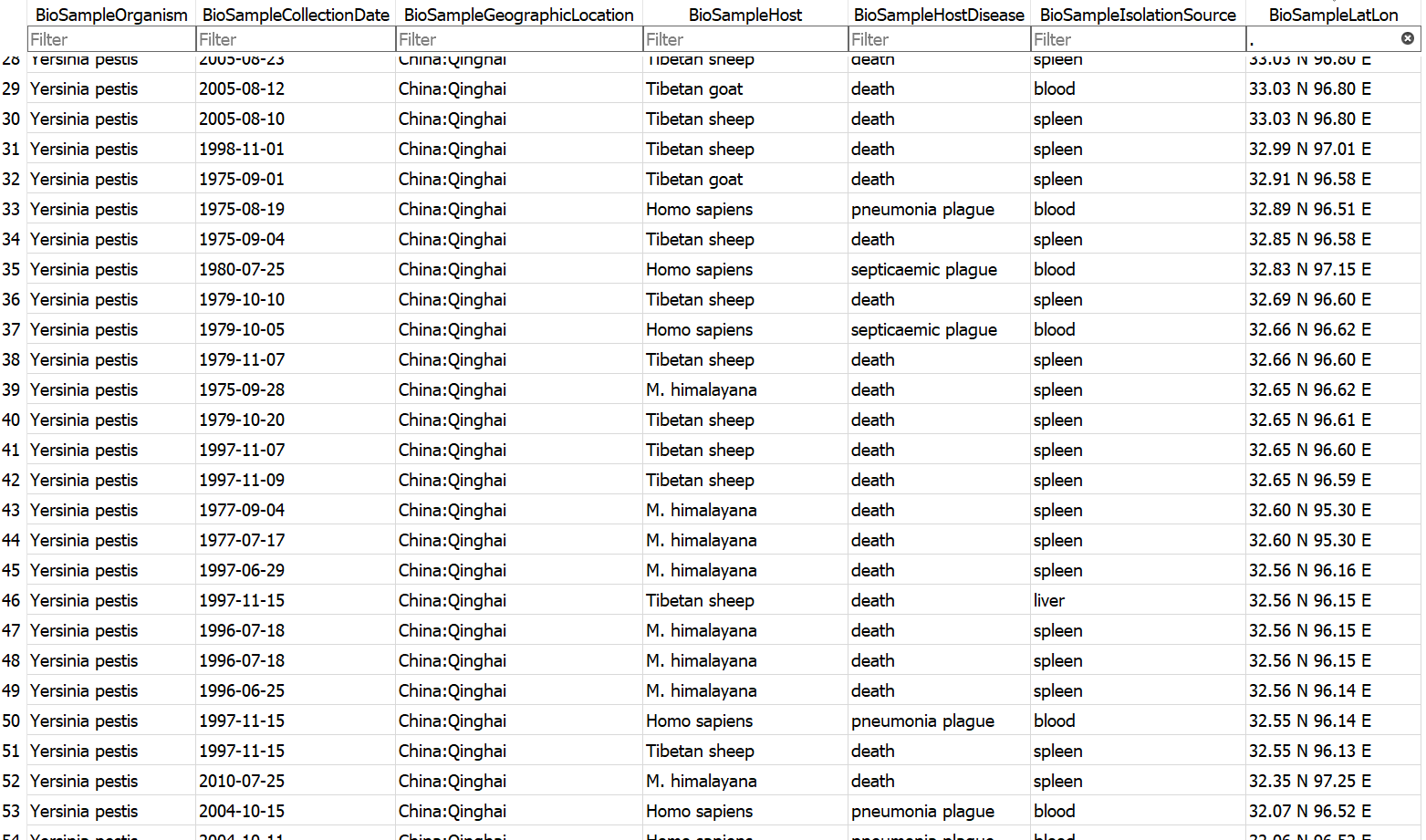
Currently Supported NCBI Tables
- Assembly
- BioProject
- BioSample
- Nucleotide
- SRA
- Pubmed
Recent and Upcoming Features
- Project "Read The Docs": Documentation Overhaul - PLANNED
- Project v0.8.3 - "Update Dependencies": Bugfixes for Installation - RELEASED
- Project v0.8.2 - "Annotate Simplicity": Simplify the Annotate Command - RELEASED
Documentation
To get started with customizing the search terms, database, and metadata fields, please read:
Issues, Questions, and Suggestions
Please submit your questions, suggestions, and bug reports to the Issue Tracker.
Please do not hesitate to post any manner of curiosity in the "Issues" tracker :) User-feedback and ideas are the most valuable resource for emerging software.
GitHub not your style? Join the NCBImeta Slack Group to see release alerts, chat with other users, and get insider perspective on development.
Contributing
Want to add features and fix bugs? Check out the Contributor's Guide for suggestions on getting started.
Citation
Eaton, K. (2020). NCBImeta: efficient and comprehensive metadata retrieval from NCBI databases. Journal of Open Source Software, 5(46), 1990, https://doi.org/10.21105/joss.01990
Authors
Author: Katherine Eaton (ktmeaton@gmail.com)
Additional Contributors
Those who have filed issues, pull-requests, and participated in discussions.
Owner
- Name: Katherine Eaton
- Login: ktmeaton
- Kind: user
- Location: Edmonton, AB
- Company: University of Alberta
- Repositories: 106
- Profile: https://github.com/ktmeaton
I am a data engineer working on the BFF-AFIRMS project: Best Future Forest -Alberta Forest Information and Genetic Resource Management Support System
JOSS Publication
NCBImeta: efficient and comprehensive metadata retrieval from NCBI databases
Authors
Tags
bioinformatics genomics metadata NCBI SRAGitHub Events
Total
- Watch event: 1
Last Year
- Watch event: 1
Committers
Last synced: 7 months ago
Top Committers
| Name | Commits | |
|---|---|---|
| Katherine Eaton | k****n@g****m | 1,051 |
| Matthew Gopez | g****m@m****a | 2 |
| Andreas Sjödin | a****n@g****m | 1 |
Committer Domains (Top 20 + Academic)
Issues and Pull Requests
Last synced: 6 months ago
All Time
- Total issues: 14
- Total pull requests: 11
- Average time to close issues: 14 days
- Average time to close pull requests: 17 days
- Total issue authors: 7
- Total pull request authors: 4
- Average comments per issue: 2.0
- Average comments per pull request: 2.45
- Merged pull requests: 8
- Bot issues: 0
- Bot pull requests: 3
Past Year
- Issues: 0
- Pull requests: 0
- Average time to close issues: N/A
- Average time to close pull requests: N/A
- Issue authors: 0
- Pull request authors: 0
- Average comments per issue: 0
- Average comments per pull request: 0
- Merged pull requests: 0
- Bot issues: 0
- Bot pull requests: 0
Top Authors
Issue Authors
- ktmeaton (7)
- druvus (2)
- hmontenegro (1)
- inesdarosa (1)
- ArchaeOphelie (1)
- evezeyl (1)
- mgopez (1)
Pull Request Authors
- ktmeaton (6)
- dependabot[bot] (3)
- mgopez (1)
- druvus (1)
Top Labels
Issue Labels
Pull Request Labels
Packages
- Total packages: 1
-
Total downloads:
- pypi 57 last-month
- Total dependent packages: 0
- Total dependent repositories: 1
- Total versions: 21
- Total maintainers: 1
pypi.org: ncbimeta
Efficient and comprehensive metadata acquisition from the NCBI databases (includes SRA).
- Homepage: https://ktmeaton.github.io/NCBImeta/
- Documentation: https://ncbimeta.readthedocs.io/
- License: MIT
-
Latest release: 0.8.3
published about 4 years ago
Rankings
Maintainers (1)
Dependencies
- ncbimeta *
- PyYAML >=5.4
- biopython >=1.74
- lxml >=4.6.3
- numpy *




