Science Score: 67.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
✓CITATION.cff file
Found CITATION.cff file -
✓codemeta.json file
Found codemeta.json file -
✓.zenodo.json file
Found .zenodo.json file -
✓DOI references
Found 4 DOI reference(s) in README -
✓Academic publication links
Links to: ncbi.nlm.nih.gov, zenodo.org -
○Committers with academic emails
-
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (12.1%) to scientific vocabulary
Keywords
Repository
Mining NCBI BLAST output
Basic Info
Statistics
- Stars: 8
- Watchers: 1
- Forks: 1
- Open Issues: 1
- Releases: 5
Topics
Metadata Files
README.md
blastMining
Mining BLAST OUTPUT
blastMining is a tool used for mining NCBI BLAST output from a single or multiple sequences,
including but not limited to ASV/OTU from amplicon sequencing,
contigs/scaffolds from shotgun metagenomics, etc.
blastMining is written in Python (tested with v3.6+). It is
available on the Python Package Index
Requirements
Before able to execute blastMining, you need to install the following programs and make sure that
they are executable and available in your PATH:
Installation
Option 1. Install via conda
This option will automatically install the dependecy programs. So, you don't need to install them manually.
bash
$ conda install -c bioconda blastmining
Option 2. Install via PyPI
bash
$ pip install blastMining
Option 3. Install manually
Download the latest realese of blastMining in my Github repository.
Then install it using pip
bash
$ pip install blastMining-1.2.0.tar.gz
Installation Notes
If you install blastMining using option 2 or option 3, you need to install the dependency programs.
You can install the dependecy programs with conda
Make sure your conda environment is up to date for the sake of the dependency programs.
```bash $ conda update -n base conda
$ conda install -c bioconda taxonkit csvtk krona blast=2.12.0
$ conda install -c conda-forge parallel ```
Before use
Don't forget to install the required databases for BLAST and TaxonKit
Tutorial
Running blastn
bash
$ blastn -query ASV.fasta \
-db nt \
-out BLASTn.out \
-outfmt="6 qseqid sseqid pident length mismatch gapopen evalue bitscore staxid" \
-max_target_seqs 10
Note: Please strict to the above blast outfmt
Next, mining your blast result with one of the following methods:
* Method A. Majority vote with percent identity cut-off
The vote algorithm is as follow:

The default percent identity cut-off is 99, 97, 95, 90, 85, 80, and 75 for Species, Genus, Family, Order, Class, Phylum, and Kingdom, respectively.
bash
$ blastMining vote \
-i BLASTn.out \
-o vote_method \
-e 0.001 \
-txl 99,97,95,90,85,80,75 \
-n 10 \
-sm 'Sample' \
-j 8 \
-p vote_method \
-kp \
-rm
* Method B. Majority vote at species level
The voteSpecies algorithm is as follow:

bash
$ blastMining voteSpecies \
-i BLASTn.out \
-o voteSpecies_method \
-e 0.001 \
-pi 99 \
-n 10 \
-sm 'Sample' \
-j 8 \
-p voteSpecies_method \
-kp \
-rm
* Method C. LCA
The lca algorithm is as follow:

The lca algorithm used in blastMining is from TaxonKit.
bash
$ blastMining lca \
-i BLASTn.out \
-o lca_method \
-e 0.001 \
-pi 95 \
-n 10 \
-sm 'Sample' \
-j 8 \
-p lca_method \
-kp \
-rm
* Method D. besthit
The besthit algorithm is as follow:
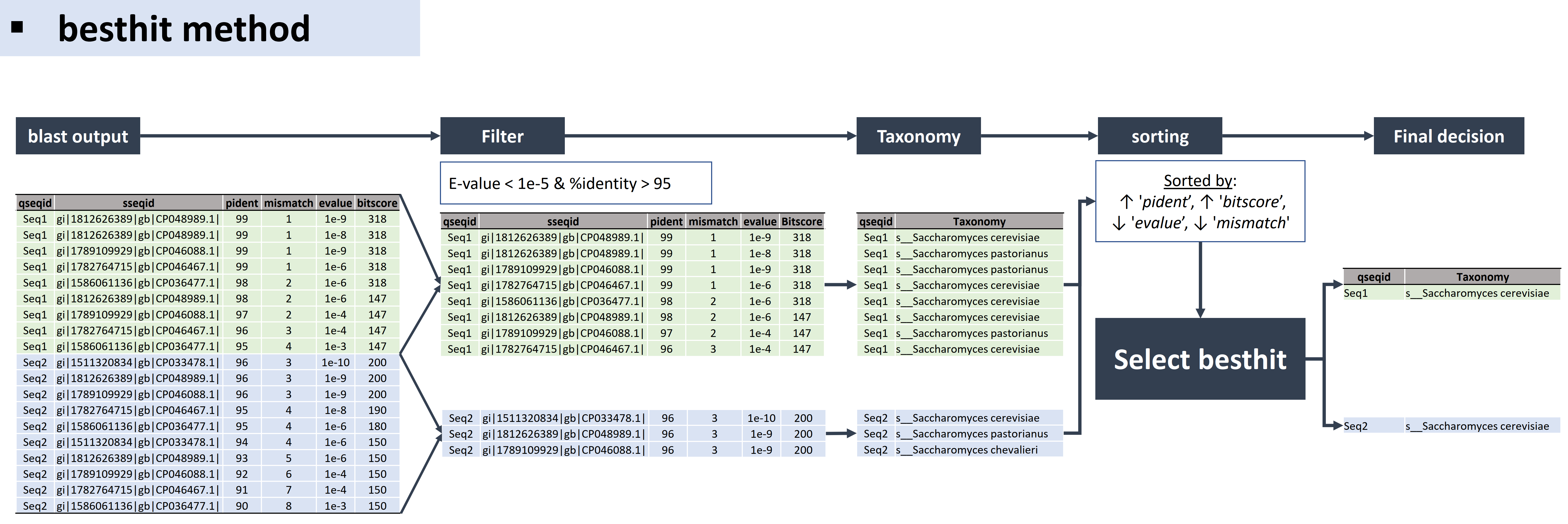
bash
$ blastMining besthit \
-i BLASTn.out \
-o besthit_method \
-e 0.001 \
-pi 97 \
-n 10 \
-sm 'Sample' \
-j 8 \
-p besthit_method \
-kp \
-rm
Full_pipeline option
This option allows you to run a full pipeline started from blastn -> blastn_output -> blastMining method -> OUTPUT.
You can select one of the following combinations:
BLAST + vote
bash
$ blastMining full_pipeline \
-i ASV.fasta \
-o vote_pipe \
-bp "-db nt -max_target_seqs 10 -num_threads 5" \
-m vote \
-e 0.001 \
-txl 99,97,95,90,85,80,75 \
-n 10 \
-sm 'Sample' \
-j 8 \
-p vote_method \
-kp \
-rm
BLAST + voteSpecies
bash
$ blastMining full_pipeline \
-i ASV.fasta \
-o voteSpecies_pipe \
-bp "-db nt -max_target_seqs 10 -num_threads 5" \
-m voteSpecies \
-e 0.001 \
-pi 99 \
-n 10 \
-sm 'Sample' \
-j 8 \
-p voteSpecies_method \
-kp \
-rm
BLAST + lca
bash
$ blastMining full_pipeline \
-i ASV.fasta \
-o lca_pipe \
-bp "-db nt -max_target_seqs 10 -num_threads 5" \
-m lca \
-e 0.001 \
-pi 99 \
-n 10 \
-sm 'Sample' \
-j 8 \
-p lca_method \
-kp \
-rm
BLAST + besthit
bash
$ blastMining full_pipeline \
-i ASV.fasta \
-o besthit_pipe \
-bp "-db nt -max_target_seqs 10 -num_threads 5" \
-m besthit \
-e 0.001 \
-pi 97 \
-n 10 \
-sm 'Sample' \
-j 8 \
-p besthit_method \
-kp \
-rm
Command options
```bash $ blastMining --help
usage: blastMining [-h] [-v] {vote,voteSpecies,lca,besthit,full_pipeline} ...
blastMining v.1.2.0
Written by: Ahmad Nuruddin Khoiri (nuruddinkhoiri34@gmail.com)
BLAST outfmt 6 only: ("qseqid","sseqid","pident","length","mismatch","gapopen","evalue","bitscore","staxid")
positional arguments: {vote,voteSpecies,lca,besthit,fullpipeline} vote blastMining: voting method with pident cut-off voteSpecies blastMining: vote at species level for all lca blastMining: lca method besthit blastMining: besthit method fullpipeline blastMining: Running BLAST + mining the output
optional arguments: -h, --help show this help message and exit -v, --version show program's version number and exit ```
Method A
```bash $ blastMining vote --help
usage: blastMining vote [-h] [-v] -i INPUT -o OUTDIR [-e EVALUE] [-txl TAXA_LEVEL] [-n TOPN] [-sm SAMPLE_NAME] [-j JOBS] [-p PREFIX] [-kp] [-rm]
blastMining: voting method with pident cut-off
blastMining v.1.2.0
Written by: Ahmad Nuruddin Khoiri (nuruddinkhoiri34@gmail.com)
optional arguments: -h, --help show this help message and exit -v, --version show program's version number and exit -i INPUT, --input INPUT blast.out file. Please use this blast outfmt 6 ONLY: ("qseqid","sseqid","pident","length","mismatch","gapopen","evalue","bitscore","staxid") [required] -o OUTDIR, --outdir OUTDIR Output directory [required] -e EVALUE, --evalue EVALUE Threshold of evalue (Ignore hits if their evalues are above this threshold) [default=1-e3] -txl TAXALEVEL, --taxalevel TAXALEVEL P.identity cut-off for Kingdom,Phylum,Class,Order,Family,Genus,Species A comma separated list of integers as an argument [default=99,97,95,90,85,80,75] -n TOPN, --topN TOPN Top N hits used for voting [default=10] -sm SAMPLENAME, --samplename SAMPLENAME Sample name in the print out table [default="sample"] -j JOBS, --jobs JOBS Number of jobs to run parallelly [default=1] -p PREFIX, --prefix PREFIX Output prefix [default='votemethod'] -kp, --kronaplot Draw krona plot [default=False] -rm, --rm_tmpdir Remove temporary directory (TMPDIR) [default=False] ```
Method B
```bash $ blastMining voteSpecies --help
usage: blastMining voteSpecies [-h] [-v] -i INPUT -o OUTDIR [-e EVALUE] [-pi PIDENT] [-n TOPN] [-sm SAMPLE_NAME] [-j JOBS] [-p PREFIX] [-kp] [-rm]
blastMining: vote at species level for all
blastMining v.1.2.0
Written by: Ahmad Nuruddin Khoiri (nuruddinkhoiri34@gmail.com)
optional arguments: -h, --help show this help message and exit -v, --version show program's version number and exit -i INPUT, --input INPUT blast.out file. Please use this blast outfmt 6 ONLY: ("qseqid","sseqid","pident","length","mismatch","gapopen","evalue","bitscore","staxid") [required] -o OUTDIR, --outdir OUTDIR Output directory [required] -e EVALUE, --evalue EVALUE Threshold of evalue (Ignore hits if their evalues are above this threshold) [default=1-e3] -pi PIDENT, --pident PIDENT Threshold of p. identity (Ignore hits if their p. identities are below this threshold) [default=99] -n TOPN, --topN TOPN Top N hits used for voting [default=10] -sm SAMPLENAME, --samplename SAMPLENAME Sample name in the print out table [default="sample"] -j JOBS, --jobs JOBS Number of jobs to run parallelly [default=1] -p PREFIX, --prefix PREFIX Output prefix [default='voteSpeciesmethod'] -kp, --kronaplot Draw krona plot [default=False] -rm, --rmtmpdir Remove temporary directory (TMPDIR) [default=False] ```
Method C
```bash $ blastMining lca --help
usage: blastMining lca [-h] [-v] -i INPUT -o OUTDIR [-e EVALUE] [-pi PIDENT] [-n TOPN] [-sm SAMPLE_NAME] [-j JOBS] [-p PREFIX] [-kp] [-rm]
blastMining: lca method
blastMining v.1.2.0
Written by: Ahmad Nuruddin Khoiri (nuruddinkhoiri34@gmail.com)
optional arguments: -h, --help show this help message and exit -v, --version show program's version number and exit -i INPUT, --input INPUT blast.out file. Please use this blast outfmt 6 ONLY: ("qseqid","sseqid","pident","length","mismatch","gapopen","evalue","bitscore","staxid") [required] -o OUTDIR, --outdir OUTDIR Output directory [required] -e EVALUE, --evalue EVALUE Threshold of evalue (Ignore hits if their evalues are above this threshold) [default=1-e3] -pi PIDENT, --pident PIDENT Threshold of p. identity (Ignore hits if their p. identities are below this threshold) [default=97] -n TOPN, --topN TOPN Top N hits used for LCA calculation [default=10] -sm SAMPLENAME, --samplename SAMPLENAME Sample name in the print out table [default="sample"] -j JOBS, --jobs JOBS Number of jobs to run parallelly [default=1] -p PREFIX, --prefix PREFIX Output prefix [default='lcamethod'] -kp, --kronaplot Draw krona plot [default=False] -rm, --rmtmpdir Remove temporary directory (TMPDIR) [default=False] ```
Method D
```bash $ blastMining besthit --help
usage: blastMining besthit [-h] [-v] -i INPUT -o OUTDIR [-e EVALUE] [-pi PIDENT] [-n TOPN] [-sm SAMPLE_NAME] [-j JOBS] [-p PREFIX] [-kp] [-rm]
blastMining: besthit method
blastMining v.1.2.0
Written by: Ahmad Nuruddin Khoiri (nuruddinkhoiri34@gmail.com)
optional arguments: -h, --help show this help message and exit -v, --version show program's version number and exit -i INPUT, --input INPUT Input file. Please use this blast outfmt 6 ONLY: ("qseqid","sseqid","pident","length","mismatch","gapopen","evalue","bitscore","staxid") [required] -o OUTDIR, --outdir OUTDIR Output directory [required] -e EVALUE, --evalue EVALUE Threshold of evalue (Ignore hits if their evalues are above this threshold) [default=1-e3] -pi PIDENT, --pident PIDENT Threshold of p. identity (Ignore hits if their p. identities are below this threshold) [default=97] -n TOPN, --topN TOPN Top N hits used for sorting [default=10] -sm SAMPLENAME, --samplename SAMPLENAME Sample name in the print out table [default="sample"] -j JOBS, --jobs JOBS Number of jobs to run parallelly [default=1] -p PREFIX, --prefix PREFIX Output prefix [default='besthitmethod'] -kp, --kronaplot Draw krona plot [default=False] -rm, --rmtmpdir Remove temporary directory (TMPDIR) [default=False] ```
Full pipeline
```bash $ blastMining full_pipeline --help
usage: blastMining fullpipeline [-h] [-v] -i INPUT -o OUTDIR -bp BLASTPARAM [-m MINING] [-e EVALUE] [-pi PIDENT] [-txl TAXA_LEVEL] [-n TOPN] [-sm SAMPLE_NAME] [-j JOBS] [-p PREFIX] [-kp] [-rm]
blastMining: Running BLAST + mining the output
blastMining v.1.2.0
Written by: Ahmad Nuruddin Khoiri (nuruddinkhoiri34@gmail.com)
optional arguments: -h, --help show this help message and exit -v, --version show program's version number and exit -i INPUT, --input INPUT input FASTA [required] -o OUTDIR, --outdir OUTDIR Output directory [required] -bp BLASTPARAM, --blastparam BLASTPARAM BLAST parameters: Note: "-outfmt" has been defined by the package, you don't need to add it [default="-db nt -numthreads 1 -maxtargetseqs 10"] -m MINING, --mining MINING blastMining method Available methods={'vote','voteSpecies','lca','besthit'} [default='vote'] -e EVALUE, --evalue EVALUE Threshold of evalue (Ignore hits if their evalues are above this threshold) [default=1-e3] -pi PIDENT, --pident PIDENT Threshold of p. identity (Ignore hits if their p. identities are below this threshold) [default=97] Required for "voteSpecies, lca, and besthit methods" Not compatible with "vote method" -txl TAXALEVEL, --taxalevel TAXALEVEL P.identity cut-off for Kingdom,Phylum,Class,Order,Family,Genus,Species [default=99,97,95,90,85,80,75] Required for "vote method" Not compatible with "voteSpecies, lca, and besthit methods" -n TOPN, --topN TOPN Top N hits used for voting [default=10] -sm SAMPLENAME, --samplename SAMPLENAME Sample name in the print out table [default="sample"] -j JOBS, --jobs JOBS Number of jobs to run parallelly [default=1] -p PREFIX, --prefix PREFIX Output prefix [default='blastMining'] -kp, --kronaplot Draw krona plot [default=False] -rm, --rmtmpdir Remove temporary directory (TMPDIR) [default=False]
```
Utility
In the case you want to convert the OUTPUT.summary to the krona-input format (OUTPUT.krona) for interactive krona pie charts visualization,
you can use the following script to do so.
bash
$ tab2krona.py -i OUTPUT.summary -o OUTPUT
The full command of the above script is as follow.
```bash
$ tab2krona.py --help
usage: tab2krona.py [-h] [-v] -i INPUT [-o OUTPUT]
convert TABLE.summary to TABLE.krona
This script is a part of blastMining program
Written by: Ahmad Nuruddin Khoiri (nuruddinkhoiri34@gmail.com)
options: -h, --help show this help message and exit -v, --version print version and exit -i INPUT, --input INPUT input table -o OUTPUT, --output OUTPUT output name [default = 'OUTPUT']
```
Citation
If you find this package useful, please cite:
BibTeX
@article{
author = {Khoiri, Ahmad Nuruddin},
title = {blastMining: mining blast output},
year = {2022},
DOI = {10.5281/zenodo.7431488},
URL = { + https://github.com/NuruddinKhoiry/blastMining},
}
Owner
- Name: Ahmad Nuruddin Khoiri
- Login: NuruddinKhoiry
- Kind: user
- Website: https://nuruddinkhoiri.blogspot.com/
- Twitter: nuruddin_khoiri
- Repositories: 3
- Profile: https://github.com/NuruddinKhoiry
PhD student at Bioinformatics and Systems Biology Program, King Mongkut's University of Technology Thonburi, Bangkok, Thailand
Citation (CITATION.cff)
blastMining: 1.2.0 message: "If you use this software, please cite it as below." authors: - family-names: "Khoiri" given-names: "Ahmad" middle-names: "Nuruddin" orcid: "https://orcid.org/0000-0003-2883-4149" title: "blastMining" version: 1.2.0 doi: 10.5281/zenodo.7431488 date-released: 2022-12-13 url: "https://github.com/NuruddinKhoiry/blastMining"
GitHub Events
Total
Last Year
Committers
Last synced: about 2 years ago
Top Committers
| Name | Commits | |
|---|---|---|
| Ahmad Nuruddin Khoiri | 3****y | 203 |
| Ahmad Nuruddin Khoiri | n****4@g****m | 1 |
Issues and Pull Requests
Last synced: 6 months ago
All Time
- Total issues: 4
- Total pull requests: 0
- Average time to close issues: 13 days
- Average time to close pull requests: N/A
- Total issue authors: 1
- Total pull request authors: 0
- Average comments per issue: 2.0
- Average comments per pull request: 0
- Merged pull requests: 0
- Bot issues: 0
- Bot pull requests: 0
Past Year
- Issues: 0
- Pull requests: 0
- Average time to close issues: N/A
- Average time to close pull requests: N/A
- Issue authors: 0
- Pull request authors: 0
- Average comments per issue: 0
- Average comments per pull request: 0
- Merged pull requests: 0
- Bot issues: 0
- Bot pull requests: 0
Top Authors
Issue Authors
- Anto007 (4)
Pull Request Authors
Top Labels
Issue Labels
Pull Request Labels
Packages
- Total packages: 1
-
Total downloads:
- pypi 19 last-month
- Total dependent packages: 0
- Total dependent repositories: 0
- Total versions: 6
- Total maintainers: 1
pypi.org: blastmining
- Homepage: https://github.com/NuruddinKhoiry/blastMining.git
- Documentation: https://blastmining.readthedocs.io/
- License: GPLv3
-
Latest release: 1.2.0
published about 3 years ago
Rankings
Maintainers (1)
Dependencies
- fastnumbers >=3.1.0
- numpy >=1.22.3
- pandas >=1.4.2
