containerit
containerit: Generating Dockerfiles for reproducible research with R - Published in JOSS (2019)
Science Score: 95.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
○CITATION.cff file
-
✓codemeta.json file
Found codemeta.json file -
✓.zenodo.json file
Found .zenodo.json file -
✓DOI references
Found 5 DOI reference(s) in README and JOSS metadata -
✓Academic publication links
Links to: joss.theoj.org -
✓Committers with academic emails
5 of 16 committers (31.3%) from academic institutions -
○Institutional organization owner
-
✓JOSS paper metadata
Published in Journal of Open Source Software
Keywords
docker
dockerfile
r
reproducible-research
reproducible-science
Keywords from Contributors
reproducibility
Last synced: 6 months ago
·
JSON representation
Repository
Package an R workspace and all dependencies as a Docker container
Basic Info
- Host: GitHub
- Owner: o2r-project
- License: gpl-3.0
- Language: R
- Default Branch: master
- Homepage: https://o2r.info/containerit/
- Size: 1.62 MB
Statistics
- Stars: 291
- Watchers: 11
- Forks: 29
- Open Issues: 86
- Releases: 2
Topics
docker
dockerfile
r
reproducible-research
reproducible-science
Created over 9 years ago
· Last pushed about 4 years ago
Metadata Files
Readme
Changelog
Contributing
License
Codemeta
README.Rmd
---
output: github_document
---
```{r knitr_config, echo = FALSE}
knitr::opts_chunk$set(
collapse = TRUE,
comment = "#>",
fig.path = "README-"
)
```
# containerit  [](https://doi.org/10.21105/joss.01603)
[](https://www.repostatus.org/#wip)
[](https://travis-ci.org/o2r-project/containerit)
[](https://ci.appveyor.com/project/nuest/containerit-rrvpq)
[](https://github.com/o2r-project/containerit/issues/68)
[](https://gitter.im/o2r-project/containerit)
`containerit` packages R script/session/workspace and all dependencies as a [Docker](https://docker.com/) container by automagically generating a suitable `Dockerfile`.
The package's website is [https://o2r.info/containerit/](https://o2r.info/containerit/).
## Prerequisites
- `containerit` only fully works if you have [Docker](https://en.wikipedia.org/wiki/Docker_(software)) installed and is only tested with [Docker Engine - Community](https://docs.docker.com/install/overview/) (previously called Docker Community Edition or Docker CE).
- `R (>= 3.5.0)` is needed so that some dependencies (e.g. BiocManager) are available; older versions of R predate the development of the package and were never tested.
## Quick start
### Try out `containerit` in a container
You can spin up a Docker container with `containerit` pre-installed if you want to try out the package.
The default of the [`containerit` images on Docker Hub](#images) is to start plain R, but you can also start an with [RStudio](https://www.rstudio.com/products/rstudio/) session in a browser.
**Note**: The `geospatial` variant is used so that examples from `containerit` vignettes are likely to work.
For a minimal `Dockerfile` to run `containerit`, see `inst/docker/minimal/Dockerfile`.
```{bash, eval=FALSE}
docker run --rm -it -e PASSWORD=o2r -p 8787:8787 o2rproject/containerit:geospatial /init
```
Now go to http://localhost:8787 and log in with the user `rstudio` and password `o2r`.
Continue in section [Use](#use).
### Install
Installation is only possible from GitHub:
```{r, eval=FALSE}
# install.packages("remotes")
remotes::install_github("o2r-project/containerit")
```
### Use
`containerit` can create `Dockerfile` objects in R and render them as `Dockerfile` instructions based on session information objects or runnable R files (`.R`, `.Rmd`).
```{r use_session}
suppressPackageStartupMessages(library("containerit"))
my_dockerfile <- containerit::dockerfile(from = utils::sessionInfo())
```
```{r use_print}
print(my_dockerfile)
```
You can disable logging:
```{r disable_logger, results='hide'}
futile.logger::flog.threshold(futile.logger::ERROR)
```
Now we create a Dockerfile for a specific R version and a given R Markdown file.
The option `filter_baseimage_pkgs` is used to not add any packages already available in the base image, which can save a lot of build time.
```{bash pull, include=FALSE}
docker pull rocker/verse:3.6.2
```
```{r use_file}
rmd_dockerfile <- containerit::dockerfile(from = "inst/demo.Rmd",
image = "rocker/verse:3.5.2",
maintainer = "o2r",
filter_baseimage_pkgs = TRUE)
print(rmd_dockerfile)
```
For extended instructions, see the vignettes at in the directory `vignettes/`, which are readable online at https://o2r.info/containerit/articles/.
## Images
```{r image_names, echo=FALSE, results='hide'}
dockerfiles_path <- "inst/docker/Dockerfile"
suppressPackageStartupMessages(library("here"))
dockerfile_latest <- readLines(here(dockerfiles_path))
base_image = sub(".*? (.+)", "\\1", dockerfile_latest[[2]])
dockerfile_geospatial <- readLines(here("inst/docker/geospatial/Dockerfile"))
geospatial_base_image = sub(".*? (.+)", "\\1", dockerfile_geospatial[[2]])
```
Images are available starting from different base images.
All images are also available with version tags.
The `Dockerfile`s are available in the directory `` `r dockerfiles_path` ``.
### verse
```{bash, eval=FALSE}
docker inspect o2rproject/containerit
```
Base image: `` `r base_image` ``
[](https://microbadger.com/images/o2rproject/containerit "Get your own version badge on microbadger.com") [](https://microbadger.com/images/o2rproject/containerit "Get your own image badge on microbadger.com") [](https://microbadger.com/images/o2rproject/containerit "Get your own commit badge on microbadger.com")
### geospatial
```{bash eval=FALSE}
docker inspect o2rproject/containerit:geospatial
```
Base image: `` `r geospatial_base_image` ``
[](https://microbadger.com/images/o2rproject/containerit:geospatial "Get your own version badge on microbadger.com") [](https://microbadger.com/images/o2rproject/containerit:geospatial "Get your own image badge on microbadger.com") [](https://microbadger.com/images/o2rproject/containerit:geospatial "Get your own commit badge on microbadger.com")
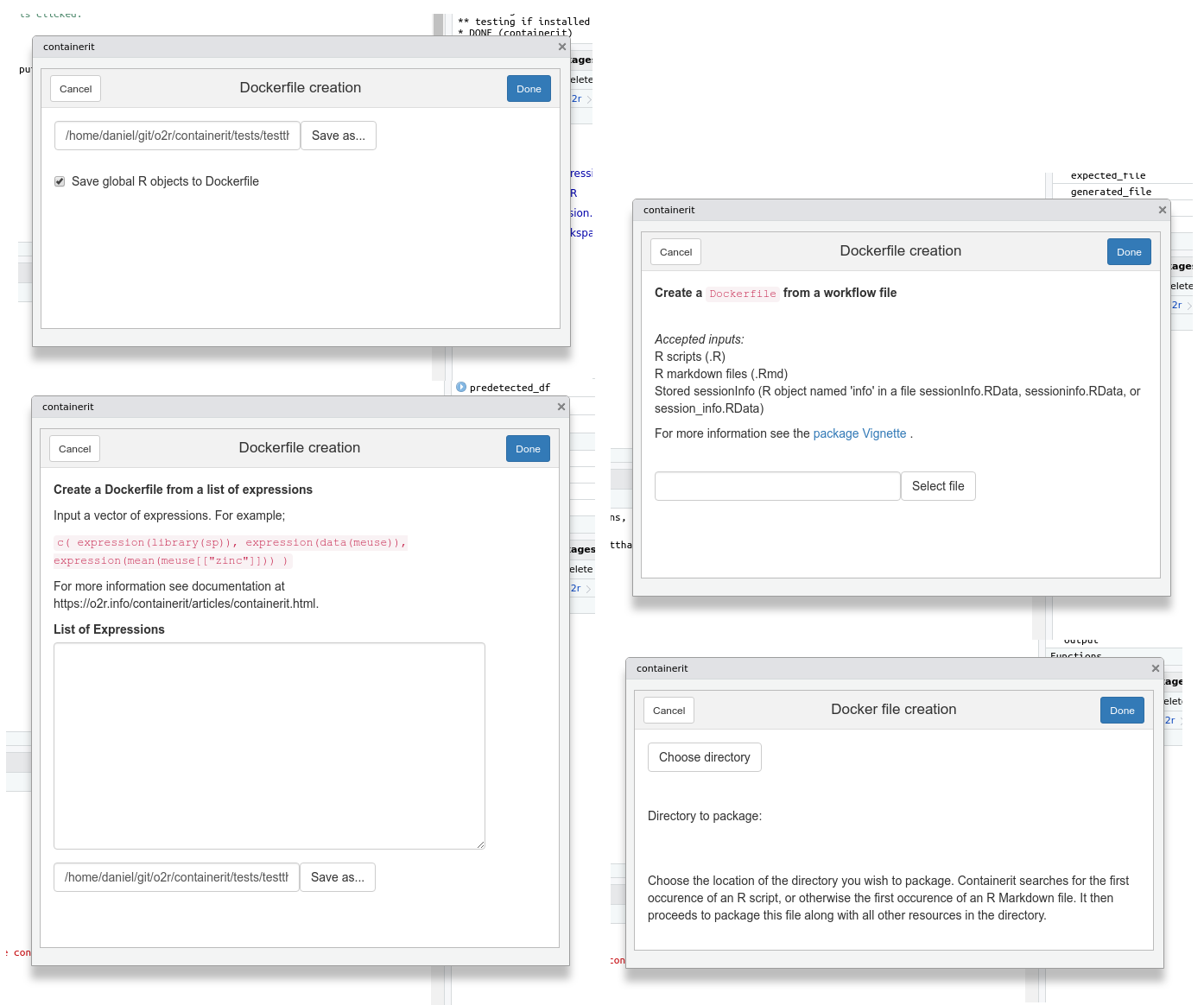
## RStudio Add-in
[RStudio Addins](https://rstudio.github.io/rstudioaddins/) allow to create interactive user interfaces for the RStudio development environment.
Courtesy of a great contribution by a [group of enthusiasts](https://github.com/o2r-project/containerit/issues/27#issuecomment-440869329) at the [ROpenSci OZ Unconference 2018](https://ozunconf18.ropensci.org/), there are several forms to quickly create `Dockefile`s from different use cases, e.g. the current session, a vector of expressions, or a script file.
## Contribute
All help is welcome: asking questions, providing documentation, testing, or even development.
Please note that this project is released with a [Contributor Code of Conduct](CONDUCT.md).
By participating in this project you agree to abide by its terms.
See [CONTRIBUTING.md](CONTRIBUTING.md) for details.
## Development
[r-hub builder](https://builder.r-hub.io/) is great for running checks, e.g. before submitting to CRAN and on other operating systems.
```{r checking, eval=FALSE}
library("rhub")
rhub::check_for_cran()
rhub::check_on_windows()
```
You can build the [`pkgdown`](https://pkgdown.r-lib.org/) site with
```{r pkgdown, eval=FALSE}
pkgdown::build_site()
```
You can build the Docker images locally with the current development version using the following commands.
```{bash dev_containers, eval=FALSE}
docker build --tag containerit:dev --file inst/docker/Dockerfile.local .
docker build --tag containerit:geospatial-dev --file inst/docker/geospatial/Dockerfile.local .
```
You can use [`pre-commit` hooks](https://github.com/lorenzwalthert/pre-commit-hooks) to avoid some mistakes.
A [codemeta](https://codemeta.github.io/) file, `codemeta.json`, with metadata about the package and its dependencies is generated automatically when this document is compiled.
```{r codemeta, results='hide'}
codemetar::write_codemeta(".")
```
## Citation
```{r citation, echo=FALSE, comment=""}
citation("containerit")
```
## License
containerit is licensed under GNU General Public License, version 3, see file LICENSE.
Copyright (C) 2019 - o2r project.
[](https://doi.org/10.21105/joss.01603)
[](https://www.repostatus.org/#wip)
[](https://travis-ci.org/o2r-project/containerit)
[](https://ci.appveyor.com/project/nuest/containerit-rrvpq)
[](https://github.com/o2r-project/containerit/issues/68)
[](https://gitter.im/o2r-project/containerit)
`containerit` packages R script/session/workspace and all dependencies as a [Docker](https://docker.com/) container by automagically generating a suitable `Dockerfile`.
The package's website is [https://o2r.info/containerit/](https://o2r.info/containerit/).
## Prerequisites
- `containerit` only fully works if you have [Docker](https://en.wikipedia.org/wiki/Docker_(software)) installed and is only tested with [Docker Engine - Community](https://docs.docker.com/install/overview/) (previously called Docker Community Edition or Docker CE).
- `R (>= 3.5.0)` is needed so that some dependencies (e.g. BiocManager) are available; older versions of R predate the development of the package and were never tested.
## Quick start
### Try out `containerit` in a container
You can spin up a Docker container with `containerit` pre-installed if you want to try out the package.
The default of the [`containerit` images on Docker Hub](#images) is to start plain R, but you can also start an with [RStudio](https://www.rstudio.com/products/rstudio/) session in a browser.
**Note**: The `geospatial` variant is used so that examples from `containerit` vignettes are likely to work.
For a minimal `Dockerfile` to run `containerit`, see `inst/docker/minimal/Dockerfile`.
```{bash, eval=FALSE}
docker run --rm -it -e PASSWORD=o2r -p 8787:8787 o2rproject/containerit:geospatial /init
```
Now go to http://localhost:8787 and log in with the user `rstudio` and password `o2r`.
Continue in section [Use](#use).
### Install
Installation is only possible from GitHub:
```{r, eval=FALSE}
# install.packages("remotes")
remotes::install_github("o2r-project/containerit")
```
### Use
`containerit` can create `Dockerfile` objects in R and render them as `Dockerfile` instructions based on session information objects or runnable R files (`.R`, `.Rmd`).
```{r use_session}
suppressPackageStartupMessages(library("containerit"))
my_dockerfile <- containerit::dockerfile(from = utils::sessionInfo())
```
```{r use_print}
print(my_dockerfile)
```
You can disable logging:
```{r disable_logger, results='hide'}
futile.logger::flog.threshold(futile.logger::ERROR)
```
Now we create a Dockerfile for a specific R version and a given R Markdown file.
The option `filter_baseimage_pkgs` is used to not add any packages already available in the base image, which can save a lot of build time.
```{bash pull, include=FALSE}
docker pull rocker/verse:3.6.2
```
```{r use_file}
rmd_dockerfile <- containerit::dockerfile(from = "inst/demo.Rmd",
image = "rocker/verse:3.5.2",
maintainer = "o2r",
filter_baseimage_pkgs = TRUE)
print(rmd_dockerfile)
```
For extended instructions, see the vignettes at in the directory `vignettes/`, which are readable online at https://o2r.info/containerit/articles/.
## Images
```{r image_names, echo=FALSE, results='hide'}
dockerfiles_path <- "inst/docker/Dockerfile"
suppressPackageStartupMessages(library("here"))
dockerfile_latest <- readLines(here(dockerfiles_path))
base_image = sub(".*? (.+)", "\\1", dockerfile_latest[[2]])
dockerfile_geospatial <- readLines(here("inst/docker/geospatial/Dockerfile"))
geospatial_base_image = sub(".*? (.+)", "\\1", dockerfile_geospatial[[2]])
```
Images are available starting from different base images.
All images are also available with version tags.
The `Dockerfile`s are available in the directory `` `r dockerfiles_path` ``.
### verse
```{bash, eval=FALSE}
docker inspect o2rproject/containerit
```
Base image: `` `r base_image` ``
[](https://microbadger.com/images/o2rproject/containerit "Get your own version badge on microbadger.com") [](https://microbadger.com/images/o2rproject/containerit "Get your own image badge on microbadger.com") [](https://microbadger.com/images/o2rproject/containerit "Get your own commit badge on microbadger.com")
### geospatial
```{bash eval=FALSE}
docker inspect o2rproject/containerit:geospatial
```
Base image: `` `r geospatial_base_image` ``
[](https://microbadger.com/images/o2rproject/containerit:geospatial "Get your own version badge on microbadger.com") [](https://microbadger.com/images/o2rproject/containerit:geospatial "Get your own image badge on microbadger.com") [](https://microbadger.com/images/o2rproject/containerit:geospatial "Get your own commit badge on microbadger.com")
## RStudio Add-in
[RStudio Addins](https://rstudio.github.io/rstudioaddins/) allow to create interactive user interfaces for the RStudio development environment.
Courtesy of a great contribution by a [group of enthusiasts](https://github.com/o2r-project/containerit/issues/27#issuecomment-440869329) at the [ROpenSci OZ Unconference 2018](https://ozunconf18.ropensci.org/), there are several forms to quickly create `Dockefile`s from different use cases, e.g. the current session, a vector of expressions, or a script file.

## Contribute
All help is welcome: asking questions, providing documentation, testing, or even development.
Please note that this project is released with a [Contributor Code of Conduct](CONDUCT.md).
By participating in this project you agree to abide by its terms.
See [CONTRIBUTING.md](CONTRIBUTING.md) for details.
## Development
[r-hub builder](https://builder.r-hub.io/) is great for running checks, e.g. before submitting to CRAN and on other operating systems.
```{r checking, eval=FALSE}
library("rhub")
rhub::check_for_cran()
rhub::check_on_windows()
```
You can build the [`pkgdown`](https://pkgdown.r-lib.org/) site with
```{r pkgdown, eval=FALSE}
pkgdown::build_site()
```
You can build the Docker images locally with the current development version using the following commands.
```{bash dev_containers, eval=FALSE}
docker build --tag containerit:dev --file inst/docker/Dockerfile.local .
docker build --tag containerit:geospatial-dev --file inst/docker/geospatial/Dockerfile.local .
```
You can use [`pre-commit` hooks](https://github.com/lorenzwalthert/pre-commit-hooks) to avoid some mistakes.
A [codemeta](https://codemeta.github.io/) file, `codemeta.json`, with metadata about the package and its dependencies is generated automatically when this document is compiled.
```{r codemeta, results='hide'}
codemetar::write_codemeta(".")
```
## Citation
```{r citation, echo=FALSE, comment=""}
citation("containerit")
```
## License
containerit is licensed under GNU General Public License, version 3, see file LICENSE.
Copyright (C) 2019 - o2r project.
Owner
- Name: Opening Reproducible Research
- Login: o2r-project
- Kind: organization
- Email: daniel.nuest@uni-muenster.de
- Location: Münster
- Website: http://o2r.info
- Repositories: 52
- Profile: https://github.com/o2r-project
JOSS Publication
containerit: Generating Dockerfiles for reproducible research with R
Published
August 21, 2019
Volume 4, Issue 40, Page 1603
Authors
Tags
containerisation Docker sandbox reproducibility reproducible researchCodeMeta (codemeta.json)
{
"@context": [
"https://doi.org/10.5063/schema/codemeta-2.0",
"http://schema.org"
],
"@type": "SoftwareSourceCode",
"identifier": "containerit",
"description": "Package R sessions, scripts, workspace directories, and R\n Markdown documents together with all dependencies to execute them in\n Docker containers. This package is supported by the project Opening\n Reproducible Research (<https://o2r.info>).",
"name": "containerit: Package R in Docker Containers",
"codeRepository": "https://github.com/o2r-project/containerit/",
"relatedLink": "https://o2r.info/containerit/",
"issueTracker": "https://github.com/o2r-project/containerit/issues",
"license": "https://spdx.org/licenses/GPL-3.0",
"version": "0.6.0.9004",
"programmingLanguage": {
"@type": "ComputerLanguage",
"name": "R",
"url": "https://r-project.org"
},
"runtimePlatform": "R version 4.1.0 (2021-05-18)",
"author": [
{
"@type": "Person",
"givenName": "Daniel",
"familyName": "Nuest",
"email": "daniel.nuest@uni-muenster.de",
"@id": "https://orcid.org/0000-0003-2392-6140"
},
{
"@type": "Person",
"givenName": "Matthias",
"familyName": "Hinz",
"email": "matthias.m.hinz@gmail.com",
"@id": "https://orcid.org/0000-0001-6837-9406"
}
],
"contributor": [
{
"@type": "Person",
"givenName": "Patrick",
"familyName": "Schratz",
"@id": "https://orcid.org/0000-0003-0748-6624"
},
{
"@type": "Person",
"givenName": "Mark",
"familyName": "Edmondson"
},
{
"@type": "Person",
"givenName": "Ben",
"familyName": "Marwick",
"@id": "https://orcid.org/0000-0001-7879-4531"
},
{
"@type": "Person",
"givenName": "Claire",
"familyName": "Miller",
"@id": "https://orcid.org/0000-0003-0758-9447"
},
{
"@type": "Person",
"givenName": "Elise",
"familyName": "Gould",
"@id": "https://orcid.org/0000-0002-6585-538X"
},
{
"@type": "Person",
"givenName": "Marc",
"familyName": "Greenaway"
}
],
"copyrightHolder": {},
"funder": {},
"maintainer": [
{
"@type": "Person",
"givenName": "Daniel",
"familyName": "Nuest",
"email": "daniel.nuest@uni-muenster.de",
"@id": "https://orcid.org/0000-0003-2392-6140"
}
],
"softwareSuggestions": [
{
"@type": "SoftwareApplication",
"identifier": "BiocGenerics",
"name": "BiocGenerics",
"provider": {
"@id": "https://www.bioconductor.org",
"@type": "Organization",
"name": "BioConductor",
"url": "https://www.bioconductor.org"
},
"sameAs": "https://bioconductor.org/packages/release/bioc/html/BiocGenerics.html"
},
{
"@type": "SoftwareApplication",
"identifier": "codetools",
"name": "codetools",
"provider": {
"@id": "https://cran.r-project.org",
"@type": "Organization",
"name": "Comprehensive R Archive Network (CRAN)",
"url": "https://cran.r-project.org"
},
"sameAs": "https://CRAN.R-project.org/package=codetools"
},
{
"@type": "SoftwareApplication",
"identifier": "covr",
"name": "covr",
"provider": {
"@id": "https://cran.r-project.org",
"@type": "Organization",
"name": "Comprehensive R Archive Network (CRAN)",
"url": "https://cran.r-project.org"
},
"sameAs": "https://CRAN.R-project.org/package=covr"
},
{
"@type": "SoftwareApplication",
"identifier": "coxrobust",
"name": "coxrobust",
"provider": {
"@id": "https://cran.r-project.org",
"@type": "Organization",
"name": "Comprehensive R Archive Network (CRAN)",
"url": "https://cran.r-project.org"
},
"sameAs": "https://CRAN.R-project.org/package=coxrobust"
},
{
"@type": "SoftwareApplication",
"identifier": "docopt",
"name": "docopt",
"provider": {
"@id": "https://cran.r-project.org",
"@type": "Organization",
"name": "Comprehensive R Archive Network (CRAN)",
"url": "https://cran.r-project.org"
},
"sameAs": "https://CRAN.R-project.org/package=docopt"
},
{
"@type": "SoftwareApplication",
"identifier": "dplyr",
"name": "dplyr",
"provider": {
"@id": "https://cran.r-project.org",
"@type": "Organization",
"name": "Comprehensive R Archive Network (CRAN)",
"url": "https://cran.r-project.org"
},
"sameAs": "https://CRAN.R-project.org/package=dplyr"
},
{
"@type": "SoftwareApplication",
"identifier": "fortunes",
"name": "fortunes",
"provider": {
"@id": "https://cran.r-project.org",
"@type": "Organization",
"name": "Comprehensive R Archive Network (CRAN)",
"url": "https://cran.r-project.org"
},
"sameAs": "https://CRAN.R-project.org/package=fortunes"
},
{
"@type": "SoftwareApplication",
"identifier": "ggplot2",
"name": "ggplot2",
"provider": {
"@id": "https://cran.r-project.org",
"@type": "Organization",
"name": "Comprehensive R Archive Network (CRAN)",
"url": "https://cran.r-project.org"
},
"sameAs": "https://CRAN.R-project.org/package=ggplot2"
},
{
"@type": "SoftwareApplication",
"identifier": "gstat",
"name": "gstat",
"provider": {
"@id": "https://cran.r-project.org",
"@type": "Organization",
"name": "Comprehensive R Archive Network (CRAN)",
"url": "https://cran.r-project.org"
},
"sameAs": "https://CRAN.R-project.org/package=gstat"
},
{
"@type": "SoftwareApplication",
"identifier": "knitr",
"name": "knitr",
"provider": {
"@id": "https://cran.r-project.org",
"@type": "Organization",
"name": "Comprehensive R Archive Network (CRAN)",
"url": "https://cran.r-project.org"
},
"sameAs": "https://CRAN.R-project.org/package=knitr"
},
{
"@type": "SoftwareApplication",
"identifier": "lattice",
"name": "lattice",
"provider": {
"@id": "https://cran.r-project.org",
"@type": "Organization",
"name": "Comprehensive R Archive Network (CRAN)",
"url": "https://cran.r-project.org"
},
"sameAs": "https://CRAN.R-project.org/package=lattice"
},
{
"@type": "SoftwareApplication",
"identifier": "lintr",
"name": "lintr",
"provider": {
"@id": "https://cran.r-project.org",
"@type": "Organization",
"name": "Comprehensive R Archive Network (CRAN)",
"url": "https://cran.r-project.org"
},
"sameAs": "https://CRAN.R-project.org/package=lintr"
},
{
"@type": "SoftwareApplication",
"identifier": "magrittr",
"name": "magrittr",
"provider": {
"@id": "https://cran.r-project.org",
"@type": "Organization",
"name": "Comprehensive R Archive Network (CRAN)",
"url": "https://cran.r-project.org"
},
"sameAs": "https://CRAN.R-project.org/package=magrittr"
},
{
"@type": "SoftwareApplication",
"identifier": "maptools",
"name": "maptools",
"provider": {
"@id": "https://cran.r-project.org",
"@type": "Organization",
"name": "Comprehensive R Archive Network (CRAN)",
"url": "https://cran.r-project.org"
},
"sameAs": "https://CRAN.R-project.org/package=maptools"
},
{
"@type": "SoftwareApplication",
"identifier": "remotes",
"name": "remotes",
"provider": {
"@id": "https://cran.r-project.org",
"@type": "Organization",
"name": "Comprehensive R Archive Network (CRAN)",
"url": "https://cran.r-project.org"
},
"sameAs": "https://CRAN.R-project.org/package=remotes"
},
{
"@type": "SoftwareApplication",
"identifier": "rgdal",
"name": "rgdal",
"provider": {
"@id": "https://cran.r-project.org",
"@type": "Organization",
"name": "Comprehensive R Archive Network (CRAN)",
"url": "https://cran.r-project.org"
},
"sameAs": "https://CRAN.R-project.org/package=rgdal"
},
{
"@type": "SoftwareApplication",
"identifier": "rmarkdown",
"name": "rmarkdown",
"provider": {
"@id": "https://cran.r-project.org",
"@type": "Organization",
"name": "Comprehensive R Archive Network (CRAN)",
"url": "https://cran.r-project.org"
},
"sameAs": "https://CRAN.R-project.org/package=rmarkdown"
},
{
"@type": "SoftwareApplication",
"identifier": "rprojroot",
"name": "rprojroot",
"provider": {
"@id": "https://cran.r-project.org",
"@type": "Organization",
"name": "Comprehensive R Archive Network (CRAN)",
"url": "https://cran.r-project.org"
},
"sameAs": "https://CRAN.R-project.org/package=rprojroot"
},
{
"@type": "SoftwareApplication",
"identifier": "sessioninfo",
"name": "sessioninfo",
"provider": {
"@id": "https://cran.r-project.org",
"@type": "Organization",
"name": "Comprehensive R Archive Network (CRAN)",
"url": "https://cran.r-project.org"
},
"sameAs": "https://CRAN.R-project.org/package=sessioninfo"
},
{
"@type": "SoftwareApplication",
"identifier": "sf",
"name": "sf",
"provider": {
"@id": "https://cran.r-project.org",
"@type": "Organization",
"name": "Comprehensive R Archive Network (CRAN)",
"url": "https://cran.r-project.org"
},
"sameAs": "https://CRAN.R-project.org/package=sf"
},
{
"@type": "SoftwareApplication",
"identifier": "sp",
"name": "sp",
"provider": {
"@id": "https://cran.r-project.org",
"@type": "Organization",
"name": "Comprehensive R Archive Network (CRAN)",
"url": "https://cran.r-project.org"
},
"sameAs": "https://CRAN.R-project.org/package=sp"
},
{
"@type": "SoftwareApplication",
"identifier": "testthat",
"name": "testthat",
"provider": {
"@id": "https://cran.r-project.org",
"@type": "Organization",
"name": "Comprehensive R Archive Network (CRAN)",
"url": "https://cran.r-project.org"
},
"sameAs": "https://CRAN.R-project.org/package=testthat"
}
],
"softwareRequirements": [
{
"@type": "SoftwareApplication",
"identifier": "R",
"name": "R",
"version": ">= 3.5.0"
},
{
"@type": "SoftwareApplication",
"identifier": "automagic",
"name": "automagic",
"provider": {
"@id": "https://cran.r-project.org",
"@type": "Organization",
"name": "Comprehensive R Archive Network (CRAN)",
"url": "https://cran.r-project.org"
},
"sameAs": "https://CRAN.R-project.org/package=automagic"
},
{
"@type": "SoftwareApplication",
"identifier": "BiocManager",
"name": "BiocManager",
"provider": {
"@id": "https://cran.r-project.org",
"@type": "Organization",
"name": "Comprehensive R Archive Network (CRAN)",
"url": "https://cran.r-project.org"
},
"sameAs": "https://CRAN.R-project.org/package=BiocManager"
},
{
"@type": "SoftwareApplication",
"identifier": "callr",
"name": "callr",
"provider": {
"@id": "https://cran.r-project.org",
"@type": "Organization",
"name": "Comprehensive R Archive Network (CRAN)",
"url": "https://cran.r-project.org"
},
"sameAs": "https://CRAN.R-project.org/package=callr"
},
{
"@type": "SoftwareApplication",
"identifier": "desc",
"name": "desc",
"provider": {
"@id": "https://cran.r-project.org",
"@type": "Organization",
"name": "Comprehensive R Archive Network (CRAN)",
"url": "https://cran.r-project.org"
},
"sameAs": "https://CRAN.R-project.org/package=desc"
},
{
"@type": "SoftwareApplication",
"identifier": "devtools",
"name": "devtools",
"provider": {
"@id": "https://cran.r-project.org",
"@type": "Organization",
"name": "Comprehensive R Archive Network (CRAN)",
"url": "https://cran.r-project.org"
},
"sameAs": "https://CRAN.R-project.org/package=devtools"
},
{
"@type": "SoftwareApplication",
"identifier": "fs",
"name": "fs",
"provider": {
"@id": "https://cran.r-project.org",
"@type": "Organization",
"name": "Comprehensive R Archive Network (CRAN)",
"url": "https://cran.r-project.org"
},
"sameAs": "https://CRAN.R-project.org/package=fs"
},
{
"@type": "SoftwareApplication",
"identifier": "futile.logger",
"name": "futile.logger",
"provider": {
"@id": "https://cran.r-project.org",
"@type": "Organization",
"name": "Comprehensive R Archive Network (CRAN)",
"url": "https://cran.r-project.org"
},
"sameAs": "https://CRAN.R-project.org/package=futile.logger"
},
{
"@type": "SoftwareApplication",
"identifier": "jsonlite",
"name": "jsonlite",
"provider": {
"@id": "https://cran.r-project.org",
"@type": "Organization",
"name": "Comprehensive R Archive Network (CRAN)",
"url": "https://cran.r-project.org"
},
"sameAs": "https://CRAN.R-project.org/package=jsonlite"
},
{
"@type": "SoftwareApplication",
"identifier": "methods",
"name": "methods"
},
{
"@type": "SoftwareApplication",
"identifier": "miniUI",
"name": "miniUI",
"provider": {
"@id": "https://cran.r-project.org",
"@type": "Organization",
"name": "Comprehensive R Archive Network (CRAN)",
"url": "https://cran.r-project.org"
},
"sameAs": "https://CRAN.R-project.org/package=miniUI"
},
{
"@type": "SoftwareApplication",
"identifier": "rjson",
"name": "rjson",
"provider": {
"@id": "https://cran.r-project.org",
"@type": "Organization",
"name": "Comprehensive R Archive Network (CRAN)",
"url": "https://cran.r-project.org"
},
"sameAs": "https://CRAN.R-project.org/package=rjson"
},
{
"@type": "SoftwareApplication",
"identifier": "rstudioapi",
"name": "rstudioapi",
"provider": {
"@id": "https://cran.r-project.org",
"@type": "Organization",
"name": "Comprehensive R Archive Network (CRAN)",
"url": "https://cran.r-project.org"
},
"sameAs": "https://CRAN.R-project.org/package=rstudioapi"
},
{
"@type": "SoftwareApplication",
"identifier": "semver",
"name": "semver",
"provider": {
"@id": "https://cran.r-project.org",
"@type": "Organization",
"name": "Comprehensive R Archive Network (CRAN)",
"url": "https://cran.r-project.org"
},
"sameAs": "https://CRAN.R-project.org/package=semver"
},
{
"@type": "SoftwareApplication",
"identifier": "shiny",
"name": "shiny",
"provider": {
"@id": "https://cran.r-project.org",
"@type": "Organization",
"name": "Comprehensive R Archive Network (CRAN)",
"url": "https://cran.r-project.org"
},
"sameAs": "https://CRAN.R-project.org/package=shiny"
},
{
"@type": "SoftwareApplication",
"identifier": "shinyFiles",
"name": "shinyFiles",
"provider": {
"@id": "https://cran.r-project.org",
"@type": "Organization",
"name": "Comprehensive R Archive Network (CRAN)",
"url": "https://cran.r-project.org"
},
"sameAs": "https://CRAN.R-project.org/package=shinyFiles"
},
{
"@type": "SoftwareApplication",
"identifier": "stevedore",
"name": "stevedore",
"provider": {
"@id": "https://cran.r-project.org",
"@type": "Organization",
"name": "Comprehensive R Archive Network (CRAN)",
"url": "https://cran.r-project.org"
},
"sameAs": "https://CRAN.R-project.org/package=stevedore"
},
{
"@type": "SoftwareApplication",
"identifier": "stringr",
"name": "stringr",
"provider": {
"@id": "https://cran.r-project.org",
"@type": "Organization",
"name": "Comprehensive R Archive Network (CRAN)",
"url": "https://cran.r-project.org"
},
"sameAs": "https://CRAN.R-project.org/package=stringr"
},
{
"@type": "SoftwareApplication",
"identifier": "sysreqs",
"name": "sysreqs",
"sameAs": "https://github.com/r-hub/sysreqs"
},
{
"@type": "SoftwareApplication",
"identifier": "utils",
"name": "utils"
},
{
"@type": "SoftwareApplication",
"identifier": "versions",
"name": "versions",
"provider": {
"@id": "https://cran.r-project.org",
"@type": "Organization",
"name": "Comprehensive R Archive Network (CRAN)",
"url": "https://cran.r-project.org"
},
"sameAs": "https://CRAN.R-project.org/package=versions"
}
],
"keywords": [
"docker",
"reproducible-research",
"reproducible-science",
"r",
"dockerfile"
],
"citation": [
{
"@type": "ScholarlyArticle",
"datePublished": "2019",
"author": [
{
"@type": "Person",
"givenName": "Daniel",
"familyName": "Nüst"
},
{
"@type": "Person",
"givenName": "Matthias",
"familyName": "Hinz"
}
],
"name": "{containerit: Generating Dockerfiles for reproducible research with R}",
"identifier": "10.21105/joss.01603",
"url": "https://doi.org/10.21105/joss.01603",
"pagination": "1603",
"@id": "https://doi.org/10.21105/joss.01603",
"sameAs": "https://doi.org/10.21105/joss.01603",
"isPartOf": {
"@type": "PublicationIssue",
"issueNumber": "40",
"datePublished": "2019",
"isPartOf": {
"@type": [
"PublicationVolume",
"Periodical"
],
"volumeNumber": "4",
"name": "{Journal of Open Source Software}"
}
}
}
],
"releaseNotes": "https://github.com/o2r-project/containerit/blob/master/NEWS.md",
"readme": "https://github.com/o2r-project/containerit/blob/master/README.md",
"fileSize": "3364.864KB",
"contIntegration": [
"https://travis-ci.org/o2r-project/containerit",
"https://ci.appveyor.com/project/nuest/containerit-rrvpq"
],
"developmentStatus": "https://www.repostatus.org/#wip"
}GitHub Events
Total
- Watch event: 3
Last Year
- Watch event: 3
Committers
Last synced: 7 months ago
Top Committers
| Name | Commits | |
|---|---|---|
| nuest | d****t@u****e | 269 |
| matthiashinz | m****z@g****m | 39 |
| Claire Miller | m****2@s****u | 10 |
| Kyle Niemeyer | k****r@g****m | 4 |
| Mark Greenaway | m****g@m****u | 4 |
| egouldo | e****d@u****u | 4 |
| Ben Marwick | b****k@h****m | 3 |
| Vanessa Sochat | v****t@s****u | 3 |
| Mark Edmondson | g****b@m****e | 2 |
| pat-s | p****z@g****m | 2 |
| Jimmy Briggs | j****1@o****m | 2 |
| 7048730 | 7****0 | 1 |
| Esther Lyon Delsordo | 5****o | 1 |
| OmaymaS | 1****S | 1 |
| MatthiasHinz | c****t@m****u | 1 |
| egouldo | e****d@g****m | 1 |
Committer Domains (Top 20 + Academic)
Issues and Pull Requests
Last synced: 6 months ago
All Time
- Total issues: 70
- Total pull requests: 30
- Average time to close issues: 7 months
- Average time to close pull requests: about 2 months
- Total issue authors: 16
- Total pull request authors: 11
- Average comments per issue: 1.36
- Average comments per pull request: 0.93
- Merged pull requests: 23
- Bot issues: 0
- Bot pull requests: 0
Past Year
- Issues: 0
- Pull requests: 0
- Average time to close issues: N/A
- Average time to close pull requests: N/A
- Issue authors: 0
- Pull request authors: 0
- Average comments per issue: 0
- Average comments per pull request: 0
- Merged pull requests: 0
- Bot issues: 0
- Bot pull requests: 0
Top Authors
Issue Authors
- nuest (48)
- MarkEdmondson1234 (3)
- vsoch (3)
- pat-s (2)
- muschellij2 (2)
- Timmimim (2)
- athammad (1)
- natemiller (1)
- cole-brokamp (1)
- holgerbrandl (1)
- mischa2002k2 (1)
- stephlocke (1)
- brnleehng (1)
- leungi (1)
- wtzhou74 (1)
Pull Request Authors
- nuest (14)
- kyleniemeyer (4)
- pat-s (2)
- benmarwick (2)
- muschellij2 (2)
- vsoch (1)
- OmaymaS (1)
- jimbrig (1)
- esther-lyondelsordo (1)
- certifiedwaif (1)
- JosiahParry (1)
Top Labels
Issue Labels
enhancement (10)
idea (7)
help wanted (5)
feature (3)
demo/documentation (2)
wontfix (1)
Pull Request Labels
Packages
- Total packages: 1
- Total downloads: unknown
- Total dependent packages: 0
- Total dependent repositories: 0
- Total versions: 2
proxy.golang.org: github.com/o2r-project/containerit
- Documentation: https://pkg.go.dev/github.com/o2r-project/containerit#section-documentation
- License: gpl-3.0
-
Latest release: v0.6.0
published over 6 years ago
Rankings
Dependent packages count: 5.5%
Average: 5.6%
Dependent repos count: 5.8%
Last synced:
6 months ago
Dependencies
DESCRIPTION
cran
- R >= 3.5.0 depends
- BiocManager * imports
- automagic * imports
- callr * imports
- desc * imports
- devtools * imports
- fs * imports
- futile.logger * imports
- jsonlite * imports
- methods * imports
- miniUI * imports
- rjson * imports
- rstudioapi * imports
- semver * imports
- shiny * imports
- shinyFiles * imports
- stevedore * imports
- stringr * imports
- sysreqs * imports
- utils * imports
- versions * imports
- BiocGenerics * suggests
- codetools * suggests
- covr * suggests
- coxrobust * suggests
- docopt * suggests
- dplyr * suggests
- fortunes * suggests
- ggplot2 * suggests
- gstat * suggests
- knitr * suggests
- lattice * suggests
- lintr * suggests
- magrittr * suggests
- maptools * suggests
- remotes * suggests
- rgdal * suggests
- rmarkdown * suggests
- rprojroot * suggests
- sessioninfo * suggests
- sf * suggests
- sp * suggests
- testthat * suggests
tests/testthat/github/DESCRIPTION
cran
- R >= 3.5.0 depends
- methods * depends
- graphics * imports
tests/testthat/package_description/DESCRIPTION
cran
- R >= 3.4.0 depends
- methods * depends
- Rcpp >= 0.12.18 imports
- grDevices * imports
- graphics * imports
- grid * imports
- units >= 0.6 imports
- utils * imports
- covr * suggests
- dplyr >= 0.8 suggests
- ggplot2 * suggests
- knitr * suggests
- lwgeom >= 0.1 suggests
- maps * suggests
