RT-utils
RT-utils: A Minimal Python Library for RT-struct Manipulation - Published in JOSS (2025)
Science Score: 95.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
○CITATION.cff file
-
✓codemeta.json file
Found codemeta.json file -
✓.zenodo.json file
Found .zenodo.json file -
✓DOI references
Found 8 DOI reference(s) in README and JOSS metadata -
✓Academic publication links
Links to: joss.theoj.org, zenodo.org -
✓Committers with academic emails
3 of 16 committers (18.8%) from academic institutions -
○Institutional organization owner
-
✓JOSS paper metadata
Published in Journal of Open Source Software
Keywords
Repository
A minimal Python library to facilitate the creation and manipulation of DICOM RTStructs.
Basic Info
Statistics
- Stars: 237
- Watchers: 9
- Forks: 70
- Open Issues: 56
- Releases: 4
Topics
Metadata Files
README.md

A minimal Python library for RT Struct manipulation
RT-Utils is motivated to allow physicians and other users to view the results of segmentation performed on a series of DICOM images. RT-Utils allows you to create or load RT Structs, extract 3d masks from RT Struct ROIs, easily add one or more regions of interest, and save the resulting RT Struct in just a few lines! You can also use the RT-Utils for merging two existing RT Structs to one file.
How it works
RT-Utils provides a builder class to faciliate the creation and loading of an RT Struct. From there, you can add ROIs through binary masks and optionally input the colour of the region along with the region name.
The format for the ROI mask is an nd numpy array of type bool. It is an array of 2d binary masks, one plane for each slice location within the DICOM series. The slices should be sorted in ascending order within the mask. Through these masks, we extract the contours of the regions of interest and place them within the RT Struct file. Note that there is currently only support for the use of one frame of reference UID and structered set ROI sequence. Also note that holes within the ROI may be handled poorly.
Installation
pip install rt_utils
libGL
libGL is a required system dependency. Install it on Linux using:
bash
sudo apt install libgl1
```
Installation in editable mode
git clone https://github.com/qurit/rt-utils.git
cd rt-utils
pip install -e .
Creating new RT Structs
```Python from rt_utils import RTStructBuilder
Create new RT Struct. Requires the DICOM series path for the RT Struct.
rtstruct = RTStructBuilder.createnew(dicomseries_path="./testlocation")
...
Create mask through means such as ML
...
Add the 3D mask as an ROI.
The colour, description, and name will be auto generated
rtstruct.addroi(mask=MASKFROMMLMODEL)
Add another ROI, this time setting the color, description, and name
rtstruct.addroi( mask=MASKFROMMLMODEL, color=[255, 0, 255], name="RT-Utils ROI!" )
rtstruct.save('new-rt-struct') ```
Adding to existing RT Structs
```Python from rt_utils import RTStructBuilder import matplotlib.pyplot as plt
Load existing RT Struct. Requires the series path and existing RT Struct path
rtstruct = RTStructBuilder.createfrom( dicomseriespath="./testlocation", rtstruct_path="./testlocation/rt-struct.dcm" )
Add ROI. This is the same as the above example.
rtstruct.addroi( mask=MASKFROMMLMODEL, color=[255, 0, 255], name="RT-Utils ROI!" )
rtstruct.save('new-rt-struct') ```
Creation Results

The results of a generated ROI with a dummy mask, as viewed in Slicer.
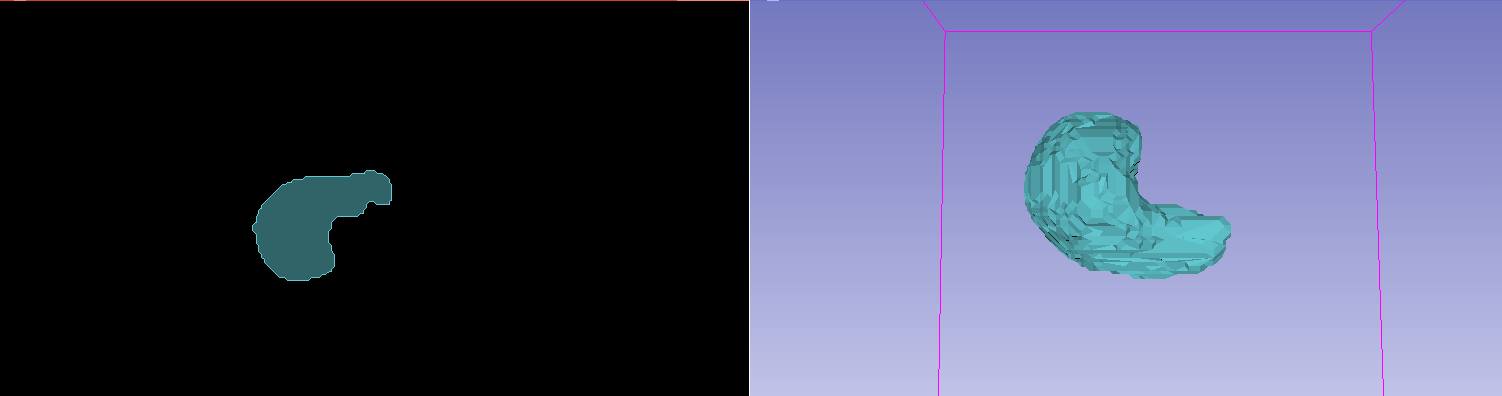
The results of a generated ROI with a liver segmentation model, as viewed in Slicer. (Note the underlying patient data has been hidden)
Loading an existing RT Struct contour as a mask
```Python from rt_utils import RTStructBuilder import matplotlib.pyplot as plt
Load existing RT Struct. Requires the series path and existing RT Struct path
rtstruct = RTStructBuilder.createfrom( dicomseriespath="./testlocation", rtstruct_path="./testlocation/rt-struct.dcm" )
View all of the ROI names from within the image
print(rtstruct.getroinames())
Loading the 3D Mask from within the RT Struct
mask3d = rtstruct.getroimaskby_name("ROI NAME")
Display one slice of the region
firstmaskslice = mask3d[:, :, 0] plt.imshow(firstmask_slice) plt.show() ```
Loading Results
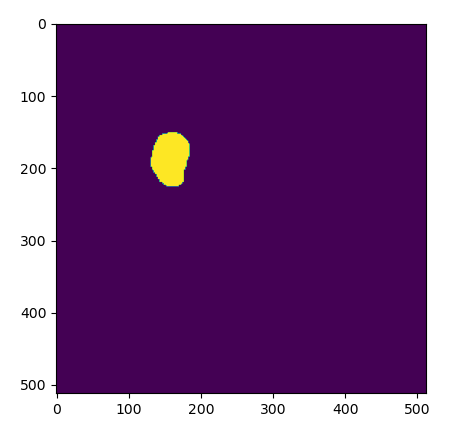
The results of a loading an exisiting ROI as a mask, as viewed in Python.
Merging two existing RT Structs
To be able to merge two RT Structs it is important that both RT Structs have to belong to the same image series, e.g. if there is one set for the organs at risk and one set for the target volume(s). ```Python from rt_utils import RTStructMerger
Load existing RT Structs and corresponding image series and merge them into one RTStruct
mergedrtstruct = RTStructMerger.mergertstructs( dicomseriespath="./testlocation", rtstructpath1="./testlocation/rt-struct1.dcm", rtstructpath2="./testlocation/rt-struct2.dcm" ) mergedrt_struct.save('merged-rt-struct') ```
Additional Parameters
The addroi method of our RTStruct class has a multitude of optional parameters available. Below is a comprehensive list of all these parameters and what they do. - color: This parameter can either be a colour string such as '#ffffff' or a RGB value as a list such as '[255, 255, 255]'. This parameter will dictate the colour of your ROI when viewed in a viewing program. If no colour is provided, RT Utils will pick from our internal colour palette based on the ROI Number of the ROI. - name: A str value that defaults to none. Used to set the name of the ROI within the RT Struct. If the name is none, RT Utils will set a name of ROI-{ROI Number}. - description: A str value that sets the description of the ROI within the RT Struct. If no value is provided, the description is just left blank. - usepinhole: A boolean value that defaults to false. If set to true, lines will be erased through your mask such that each separate region within your image can be encapsulated via a single contour instead of contours nested within one another. Use this if your RT Struct viewer of choice does not support nested contours / contours with holes. - approximatecontours: A boolean value that defaults to True which defines whether or not approximations are made when extracting contours from the input mask. Setting this to false will lead to much larger contour data within your RT Struct so only use this if as much precision as possible is required. - roigenerationalgorithm: An enum value that defaults to 0 which defines what ROI generation algorithm will be used. 0=\'AUTOMATIC\', 1=\'SEMIAUTOMATIC\', or 2=\'MANUAL\'.
🚀 New features
- nifti to rtstruct conversion has been added.
- conversion from DICOM to NIFTI for PET and corresponding RT-struct file has been added as an example usage with no issues of shift or mismatch between the masks.
Examples
You can find the comparison between RT-utils and other conversion techniques in this public repository: https://github.com/qurit/dicomnifticonversion_project
Examples
You can find the comparison between RT-utils and other conversion techniques in this public repository: https://github.com/qurit/dicomnifticonversion_project.
The real-world example in the JOSS paper under review was generated using the code provided in the RTUTILSreadworldexamplefor_PET.ipynb.
Contributing
We welcome contributions to this project! Please review our CONTRIBUTING guidelines for more details.
How to Cite
If you are incorporating RT-Utils into your projects, kindly include the following citation:
Read the full paper: Asim Shrestha, Adam Watkins, Fereshteh Yousefirizi, Arman Rahmim, and Carlos Uribe RT-utils: A Minimal Python Library for RT-struct Manipulation
Owner
- Name: Qurit
- Login: qurit
- Kind: organization
- Location: Vancouver
- Website: https://qurit.ca
- Twitter: CarlosUribeM
- Repositories: 17
- Profile: https://github.com/qurit
Quantitative Radiomolecular Imaging & Therapy Lab
JOSS Publication
RT-utils: A Minimal Python Library for RT-struct Manipulation
Authors
Department of Integrative Oncology, BC Cancer Research Institute, Vancouver, British Columbia, Canada
Department of Integrative Oncology, BC Cancer Research Institute, Vancouver, British Columbia, Canada

Department of Integrative Oncology, BC Cancer Research Institute, Vancouver, British Columbia, Canada

Department of Integrative Oncology, BC Cancer Research Institute, Vancouver, British Columbia, Canada, Department of Radiology, University of British Columbia, Vancouver, BC, Canada, Departments of Physics and Biomedical Engineering, University of British Columbia, Vancouver, BC, Canada, Department of Biomedical Engineering, University of British Columbia, Vancouver, Canada
Tags
RT-struct Radiotherapy structures mask DICOM image segmentationGitHub Events
Total
- Create event: 4
- Release event: 1
- Issues event: 3
- Watch event: 35
- Delete event: 5
- Issue comment event: 22
- Push event: 63
- Pull request review event: 7
- Pull request event: 16
- Fork event: 14
Last Year
- Create event: 4
- Release event: 1
- Issues event: 3
- Watch event: 35
- Delete event: 5
- Issue comment event: 22
- Push event: 63
- Pull request review event: 7
- Pull request event: 16
- Fork event: 14
Committers
Last synced: 7 months ago
Top Committers
| Name | Commits | |
|---|---|---|
| Asim Shrestha | a****h@s****a | 122 |
| Fereshteh Yousefirizi | 8****i | 92 |
| Adam Watkins | 3****s | 36 |
| Zhack47 | 5****7 | 9 |
| ThomasBudd | t****8@c****k | 4 |
| Adam Zyzik | a****k@g****m | 3 |
| Pedro Esquinas | p****i@g****m | 3 |
| Carlos F. Uribe | c****i | 2 |
| Pedro | p****o@p****m | 2 |
| Robin Hegering | 4****9 | 2 |
| igorhlx | i****r@c****i | 2 |
| Mathieu Goulet | m****5@g****m | 1 |
| Maxence Larose | m****1@u****a | 1 |
| Phillip Chlap | p****p@u****u | 1 |
| Samuel Ouellet | s****0@u****a | 1 |
| Tom Roberts | t****s@k****k | 1 |
Committer Domains (Top 20 + Academic)
Issues and Pull Requests
Last synced: 6 months ago
All Time
- Total issues: 70
- Total pull requests: 69
- Average time to close issues: about 1 month
- Average time to close pull requests: about 1 month
- Total issue authors: 53
- Total pull request authors: 27
- Average comments per issue: 2.01
- Average comments per pull request: 0.87
- Merged pull requests: 38
- Bot issues: 0
- Bot pull requests: 0
Past Year
- Issues: 3
- Pull requests: 17
- Average time to close issues: N/A
- Average time to close pull requests: about 1 month
- Issue authors: 3
- Pull request authors: 2
- Average comments per issue: 0.33
- Average comments per pull request: 0.12
- Merged pull requests: 7
- Bot issues: 0
- Bot pull requests: 0
Top Authors
Issue Authors
- asim-shrestha (8)
- awtkns (4)
- ThomasBudd (2)
- jrenslo (2)
- 1413194910 (2)
- IlvaHou55 (2)
- Suman-c21 (2)
- mathiser (2)
- Zhack47 (2)
- tomaroberts (2)
- FabienRech (2)
- Kimhasen (1)
- MaziarSabouri (1)
- nadKo375 (1)
- TmnGitHub (1)
Pull Request Authors
- qurit-frizi (25)
- asim-shrestha (10)
- awtkns (7)
- gacou54 (4)
- rheg49 (3)
- Zhack47 (2)
- VendenIX (2)
- MLK97 (2)
- tomaroberts (1)
- smichi23 (1)
- goma1256 (1)
- FabienRech (1)
- Brian-Taylor8 (1)
- jrenslo (1)
- plesqui (1)
Top Labels
Issue Labels
Pull Request Labels
Dependencies
- dataclasses *
- numpy *
- opencv-python *
- pydicom ==2.1.2
- psf/black stable composite
- actions/checkout v2 composite
- actions/setup-python v2 composite