aggmap
Jigsaw-like AggMap: A Robust and Explainable Multi-Channel Omics Deep Learning Tool
Science Score: 36.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
○CITATION.cff file
-
✓codemeta.json file
Found codemeta.json file -
✓.zenodo.json file
Found .zenodo.json file -
○DOI references
-
○Academic publication links
-
✓Committers with academic emails
1 of 7 committers (14.3%) from academic institutions -
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (11.2%) to scientific vocabulary
Keywords
Keywords from Contributors
Repository
Jigsaw-like AggMap: A Robust and Explainable Multi-Channel Omics Deep Learning Tool
Basic Info
- Host: GitHub
- Owner: shenwanxiang
- License: gpl-3.0
- Language: Jupyter Notebook
- Default Branch: master
- Homepage: https://bidd-aggmap.readthedocs.io/en/latest/
- Size: 871 MB
Statistics
- Stars: 34
- Watchers: 1
- Forks: 5
- Open Issues: 2
- Releases: 4
Topics
Metadata Files
README.md

Jigsaw-like AggMap
A Robust and Explainable Omics Deep Learning Tool
Installation (Only on Linux system)
install aggmap by: ```bash
create an aggmap env
conda create -n aggmap python=3.8 conda activate aggmap pip install --upgrade pip pip install aggmap==1.2.1 ```
Usage
```python import pandas as pd from sklearn.datasets import loadbreastcancer from aggmap import AggMap, AggMapNet
Data loading
data = loadbreastcancer() dfx = pd.DataFrame(data.data, columns=data.featurenames) dfy = pd.getdummies(pd.Series(data.target))
AggMap object definition, fitting, and saving
mp = AggMap(dfx, metric = 'correlation') mp.fit(clusterchannels=5, embmethod = 'umap', verbose=0) mp.save('agg.mp')
AggMap visulizations: Hierarchical tree, embeddng scatter and grid
mp.plottree() mp.plotscatter(enableddatalabels=True, radius=5) mp.plotgrid(enableddata_labels=True)
Transoformation of 1d vectors to 3D Fmaps (-1, w, h, c) by AggMap
X = mp.batchtransform(dfx.values, njobs=4, scale_method = 'minmax') y = dfy.values
AggMapNet training, validation, early stopping, and saving
clf = AggMapNet.MultiClassEstimator(epochs=50, gpuid=0) clf.fit(X, y, Xvalid=None, yvalid=None) clf.save_model('agg.model')
Model explaination by simply-explainer: global, local
simpexplainer = AggMapNet.simplyexplainer(clf, mp) globalsimpimportance = simpexplainer.globalexplain(clf.X, clf.y) localsimpimportance = simpexplainer.localexplain(clf.X[[0]], clf.y[[0]])
Model explaination by shapley-explainer: global, local
shapexplainer = AggMapNet.shapleyexplainer(clf, mp) globalshapimportance = shapexplainer.globalexplain(clf.X) localshapimportance = shapexplainer.localexplain(clf.X[[0]]) ```
How It Works?
- AggMap flowchart of feature mapping and agglomeration into ordered (spatially correlated) multi-channel feature maps (Fmaps)
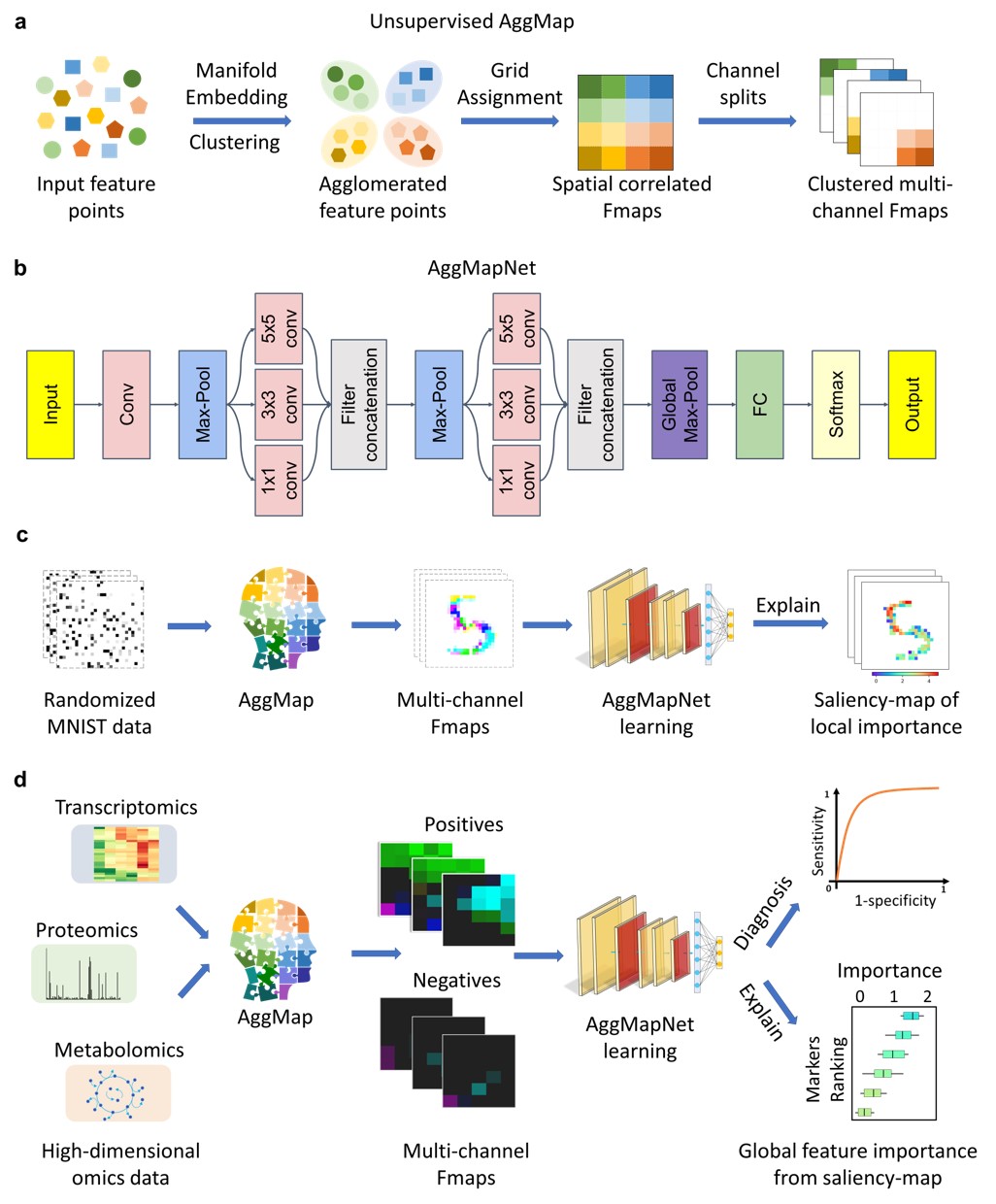 a, AggMap flowchart of feature mapping and aggregation into ordered (spatially-correlated) channel-split feature maps (Fmaps).b, CNN-based AggMapNet architecture for Fmaps learning. c, proof-of-concept illustration of AggMap restructuring of unordered data (randomized MNIST) into clustered channel-split Fmaps (reconstructed MNIST) for CNN-based learning and important feature analysis. d, typical biomedical applications of AggMap in restructuring omics data into channel-split Fmaps for multi-channel CNN-based diagnosis and biomarker discovery (explanation
a, AggMap flowchart of feature mapping and aggregation into ordered (spatially-correlated) channel-split feature maps (Fmaps).b, CNN-based AggMapNet architecture for Fmaps learning. c, proof-of-concept illustration of AggMap restructuring of unordered data (randomized MNIST) into clustered channel-split Fmaps (reconstructed MNIST) for CNN-based learning and important feature analysis. d, typical biomedical applications of AggMap in restructuring omics data into channel-split Fmaps for multi-channel CNN-based diagnosis and biomarker discovery (explanation saliency-map of important features).
Proof-of-Concepts of reconstruction ability on MNIST Dataset
- It can reconstruct to the original image from completely randomly permuted (disrupted) MNIST data:
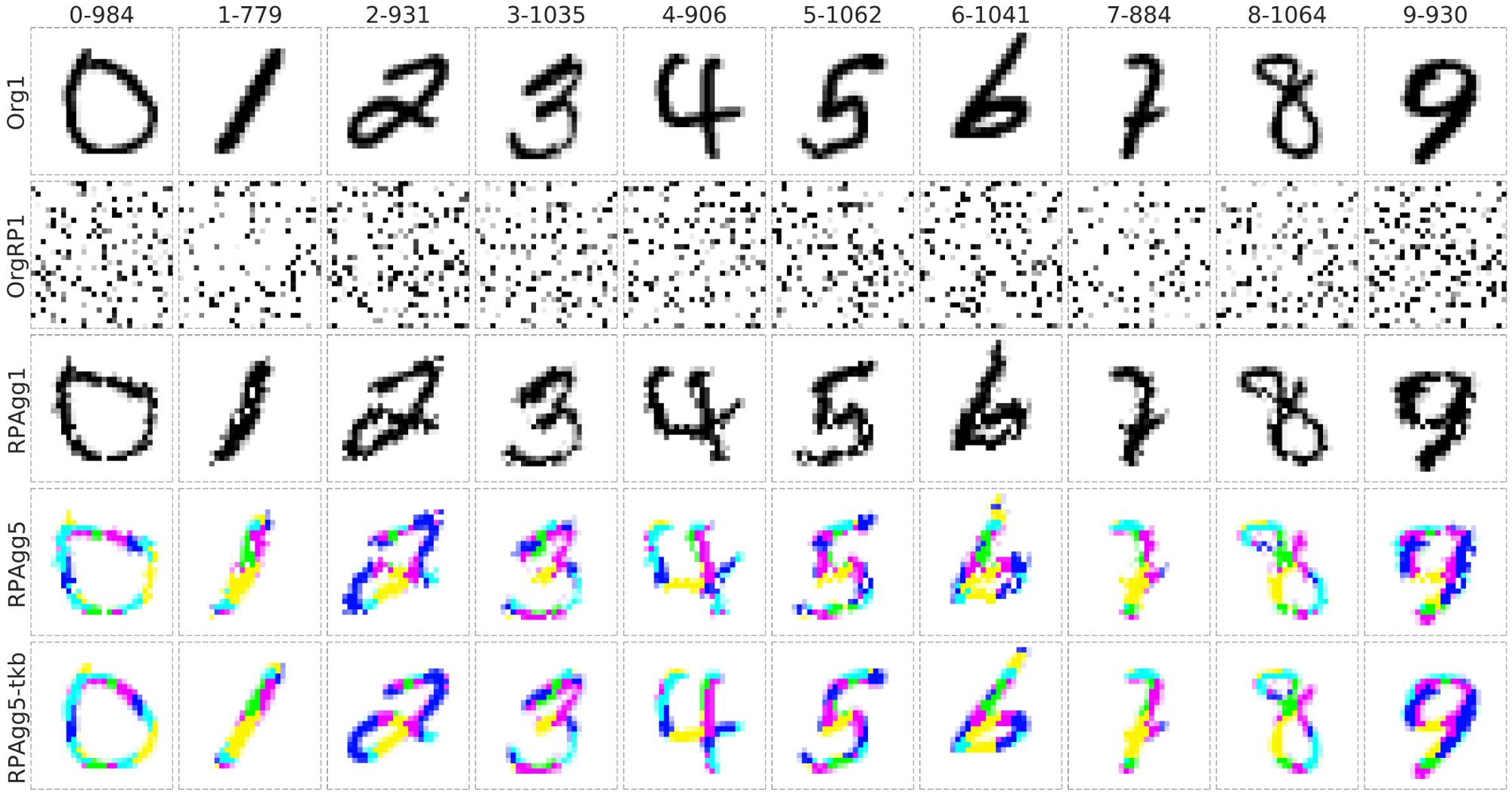
Org1: the original grayscale images (channel = 1), OrgRP1: the randomized images of Org1 (channel = 1), RPAgg1, 5: the reconstructed images of OrgPR1 by AggMap feature restructuring (channel = 1, 5 respectively, each color represents features of one channel). RPAgg5-tkb: the original images with the pixels divided into 5 groups according to the 5-channels of RPAgg5 and colored in the same way as RPAgg5.
The effect of the number of channels on model performance
- Multi-channel Fmaps can boost the model performance notably:
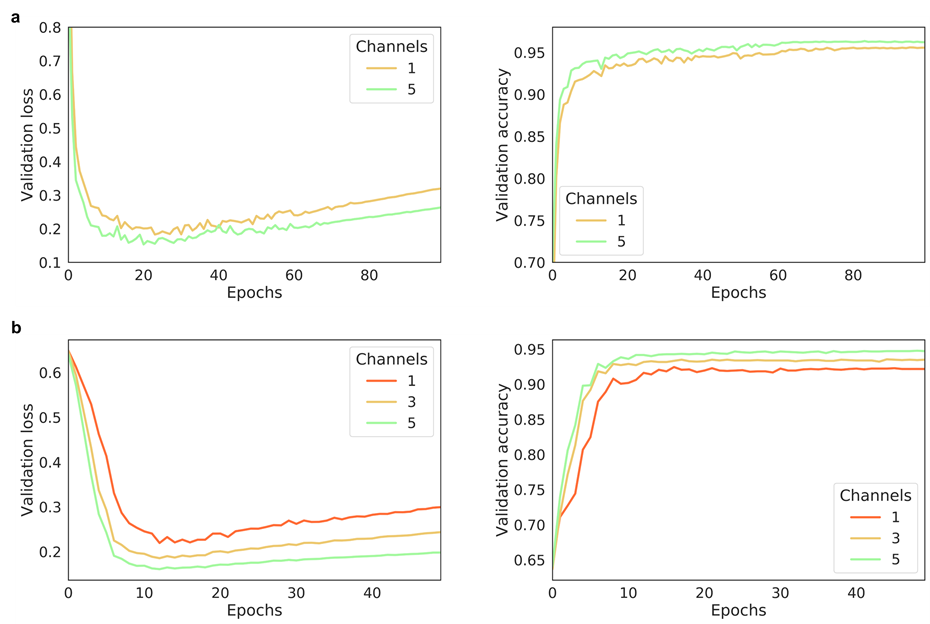
The performance of AggMapNet using different number of channels on the TCGA-T (a) and COV-D (b). For TCGA-T, ten-fold cross validation average performance, for COV-D, a fivefold cross validation was performed and repeat 5 rounds using different random seeds (total 25 training times), their average performances of the validation set were reported.
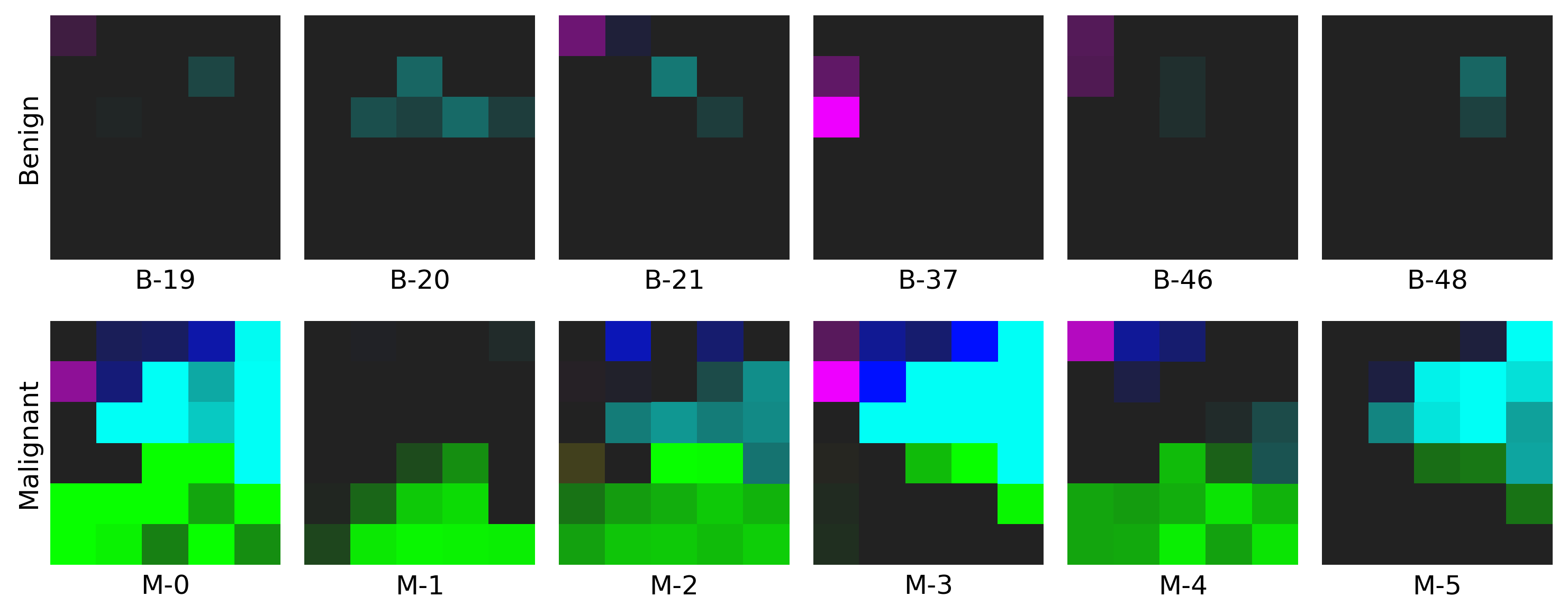
Example for Restructured Fmaps
- The example on WDBC dataset: click here to find out more!
Citation
Shen, Wan Xiang, et al. "AggMapNet: enhanced and explainable low-sample omics deep learning with feature-aggregated multi-channel networks." Nucleic Acids Research 50.8 (2022): e45-e45.
Owner
- Name: Charleshen
- Login: shenwanxiang
- Kind: user
- Location: Singapore
- Company: FoS, National University of Singapore
- Website: www.shenwx.com
- Twitter: WanXiang_Shen
- Repositories: 6
- Profile: https://github.com/shenwanxiang
A data scientist, interested in Bioinformatics & Chemoinformatics
GitHub Events
Total
- Watch event: 4
- Fork event: 2
Last Year
- Watch event: 4
- Fork event: 2
Committers
Last synced: over 1 year ago
Top Committers
| Name | Commits | |
|---|---|---|
| shenwanxiang | s****g@t****n | 216 |
| Charleshen | s****g@1****m | 86 |
| shenwanxiang | s****g@t****n | 61 |
| shenwanxiang | s****g@g****m | 7 |
| shenwanxiang | h****g@g****m | 5 |
| Shen | w****6@c****u | 1 |
| dependabot[bot] | 4****] | 1 |
Committer Domains (Top 20 + Academic)
Issues and Pull Requests
Last synced: over 1 year ago
All Time
- Total issues: 6
- Total pull requests: 12
- Average time to close issues: 13 days
- Average time to close pull requests: about 2 months
- Total issue authors: 5
- Total pull request authors: 2
- Average comments per issue: 2.17
- Average comments per pull request: 0.67
- Merged pull requests: 3
- Bot issues: 0
- Bot pull requests: 9
Past Year
- Issues: 0
- Pull requests: 0
- Average time to close issues: N/A
- Average time to close pull requests: N/A
- Issue authors: 0
- Pull request authors: 0
- Average comments per issue: 0
- Average comments per pull request: 0
- Merged pull requests: 0
- Bot issues: 0
- Bot pull requests: 0
Top Authors
Issue Authors
- shenwanxiang (2)
- computbiol (1)
- ivy-yuan (1)
- tttender (1)
- dxbdxx (1)
Pull Request Authors
- dependabot[bot] (9)
- shenwanxiang (3)
Top Labels
Issue Labels
Pull Request Labels
Packages
- Total packages: 1
-
Total downloads:
- pypi 121 last-month
- Total dependent packages: 0
- Total dependent repositories: 1
- Total versions: 10
- Total maintainers: 1
pypi.org: aggmap
Jigsaw-like AggMap: A Robust and Explainable Omics Deep Learning Tool
- Homepage: https://github.com/shenwanxiang/bidd-aggmap/tree/master
- Documentation: https://aggmap.readthedocs.io/
- License: gpl-3.0
-
Latest release: 1.2.1
published over 2 years ago
Rankings
Maintainers (1)
Dependencies
- IPython *
- Sphinx ==5.1.1
- bokeh *
- ipykernel *
- nbsphinx *
- sphinx-rtd-theme *
- sphinx_gallery *

