Af-analysis
Af-analysis: a Python package for Alphafold analysis - Published in JOSS (2025)
Science Score: 100.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
✓CITATION.cff file
Found CITATION.cff file -
✓codemeta.json file
Found codemeta.json file -
✓.zenodo.json file
Found .zenodo.json file -
✓DOI references
Found 40 DOI reference(s) in README and JOSS metadata -
✓Academic publication links
Links to: biorxiv.org, nature.com, joss.theoj.org, zenodo.org -
✓Committers with academic emails
1 of 2 committers (50.0%) from academic institutions -
○Institutional organization owner
-
✓JOSS paper metadata
Published in Journal of Open Source Software
Scientific Fields
Repository
Analysis of alphafold and colabfold results
Basic Info
- Host: GitHub
- Owner: samuelmurail
- License: gpl-2.0
- Language: Jupyter Notebook
- Default Branch: main
- Size: 51.2 MB
Statistics
- Stars: 31
- Watchers: 2
- Forks: 6
- Open Issues: 3
- Releases: 1
Metadata Files
README.md
About Alphafold Analysis

af-analysis is a python package for the analysis of AlphaFold protein structure predictions.
This package is designed to simplify and streamline the process of working with protein structures
generated by:
Source code repository: https://github.com/samuelmurail/af_analysis
Statement of Need
AlphaFold 2 and its derivatives have revolutionized protein structure prediction, achieving remarkable accuracy.
Analyzing the abundance of resulting structural models can be challenging and time-consuming.
Existing tools often require separate scripts for calculating various quality metrics (pDockQ, pDockQ2, LIS score) and assessing model diversity.
af-analysis addresses these challenges by providing a unified and user-friendly framework for in-depth analysis of AlphaFold 2 results.
Main features
- Import AlphaFold or ColabFold prediction directories as pandas DataFrames for efficient data handling.
- Calculate and add additional structural quality metrics to the DataFrame, including:
- pDockQ
- pDockQ2
- LIS score
- Visualize predicted protein models.
- Cluster generated models to identify diverse conformations.
- Select the best models based on defined criteria.
- Add your custom metrics to the DataFrame for further analysis.
Installation
af-analysisis available on PyPI and can be installed usingpip:
bash
pip install af_analysis
- You can install last version from the github repo:
bash
pip install git+https://github.com/samuelmurail/af_analysis.git@main
- AF-Analysis can also be installed easily through github:
bash
git clone https://github.com/samuelmurail/af_analysis
cd af_analysis
pip install .
Documentation
The complete documentation is available at ReadTheDocs.
A notebook showing the basic usage of the
af_analysislibrary can be found here.Alternatively you can test is directly on Google colab:
Usage
Importing data
Create the Data object, giving the path of the directory containing the results of the alphafold2/colabfold run.
python
import af_analysis
my_data = af_analysis.Data('MY_AF_RESULTS_DIR')
Extracted data are available in the df attribute of the Data object.
python
my_data.df
Analysis
python
from af_analysis import analysis
analysis.pdockq(my_data)
analysis.pdockq2(my_data)
Docking Analysis
- The
dockingpackage contains several function to add metrics like LIS Score:
python
from af_analysis import docking
docking.LIS_pep(my_data)
Plots
- At first approach the user can visualize the pLDDT, PAE matrix and the model scores. The
show_info()function displays the scores of the models, as well as the pLDDT plot and PAE matrix in a interactive way.
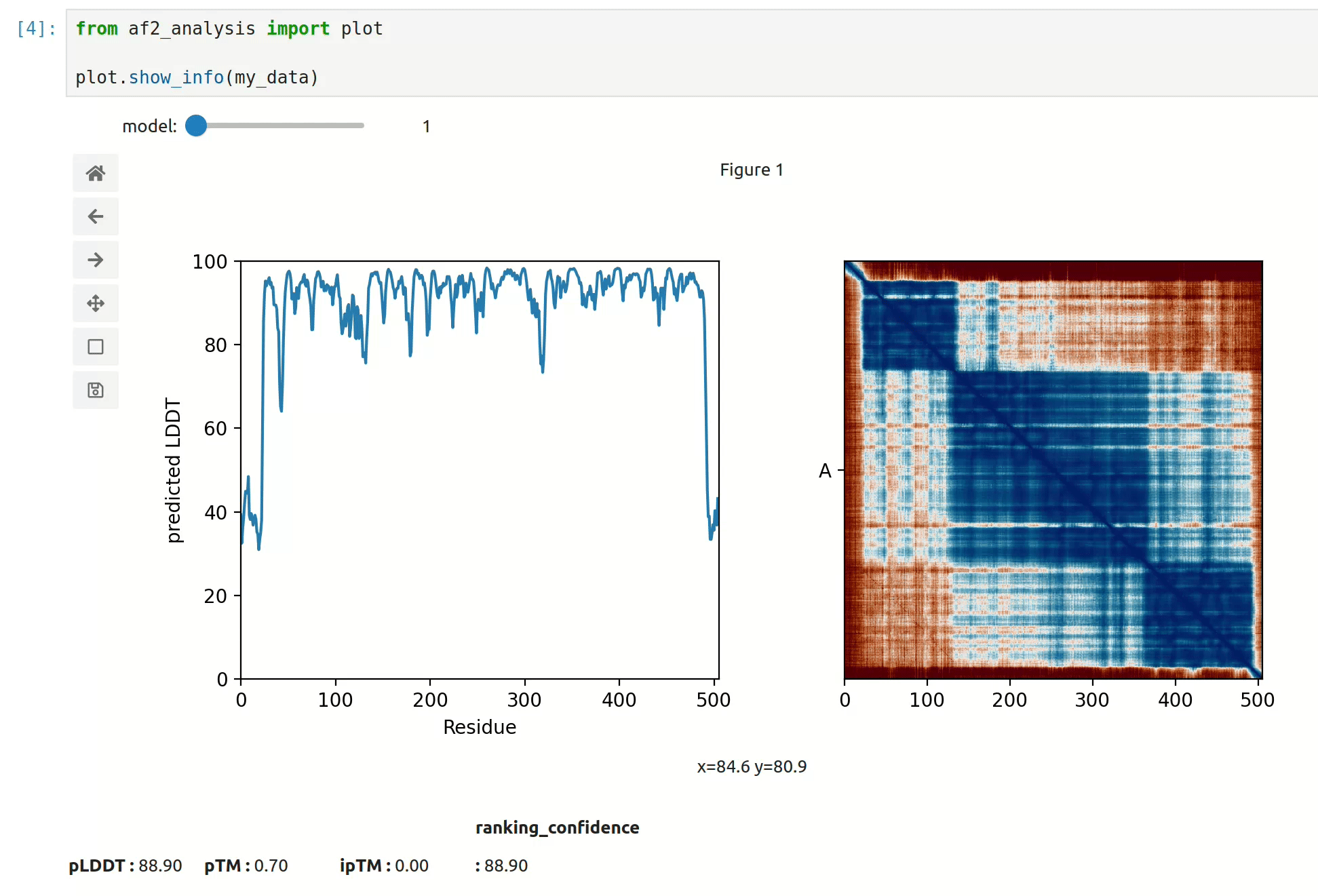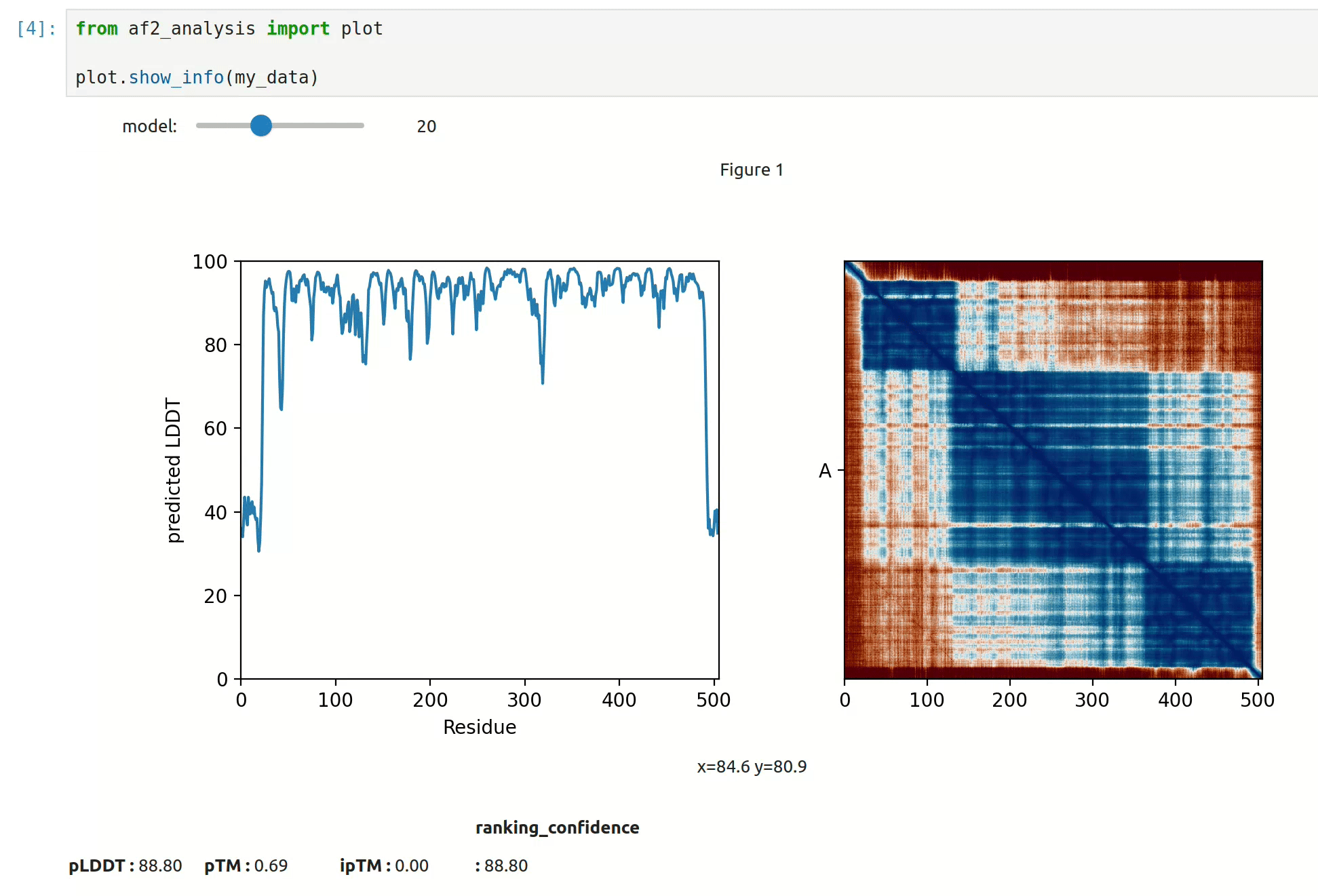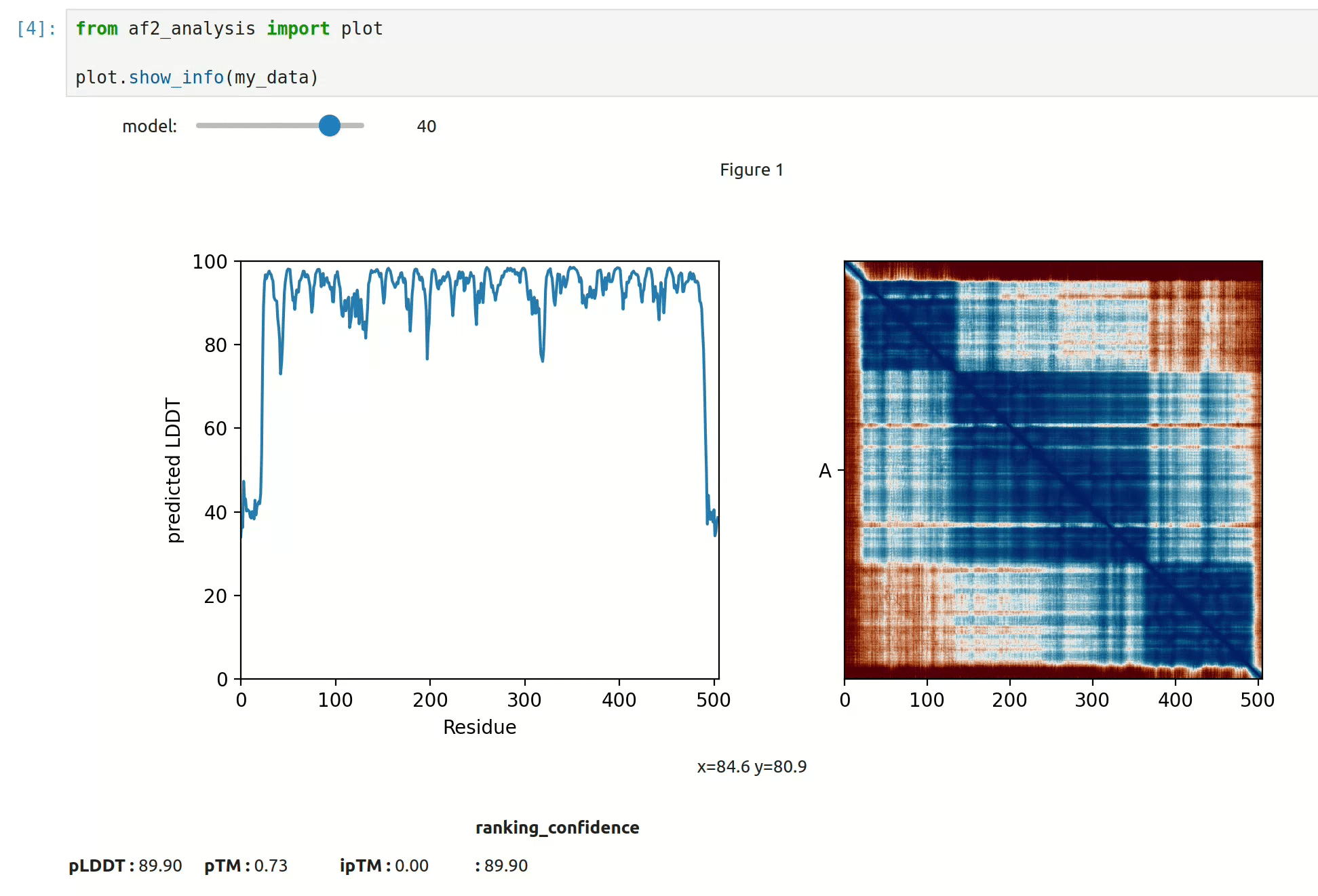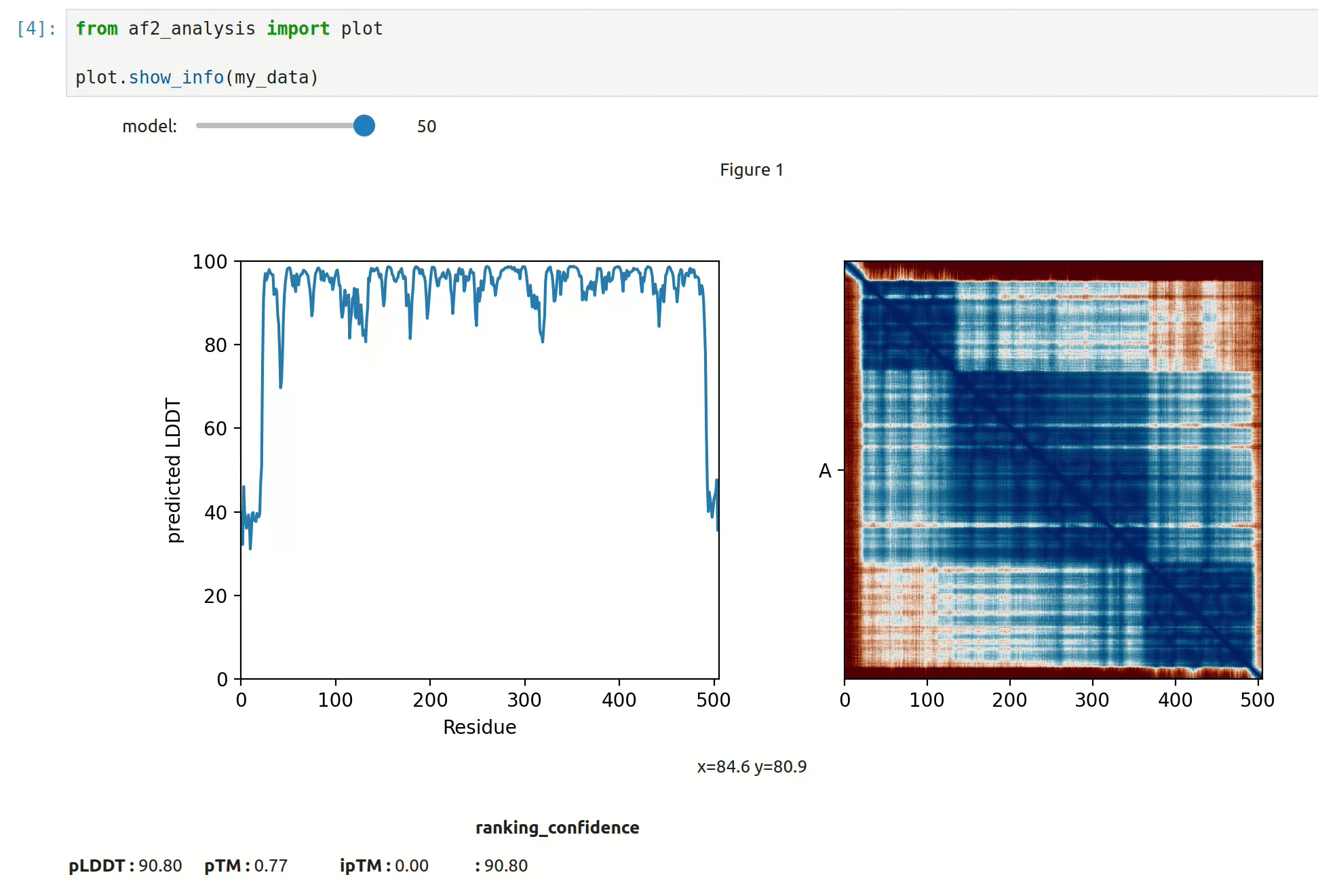
- plot msa, plddt and PAE:
python
my_data.plot_msa()
my_data.plot_plddt([0,1])
best_model_index = my_data.df['ranking_confidence'].idxmax()
my_data.plot_pae(best_model_index)
- show 3D structure (
nglviewpackage required):
python
my_data.show_3d(my_data.df['ranking_confidence'].idxmax())
Dependencies
af_analysis requires the following dependencies:
pdb_numpypandasnumpytqdmseaborncmcrameringlviewipywidgetsmdanalysis
Contributing
af-analysis is an open-source project and contributions are welcome. If
you find a bug or have a feature request, please open an issue on the GitHub
repository at https://github.com/samuelmurail/af_analysis. If you would like
to contribute code, please fork the repository and submit a pull request.
Authors
- Alaa Regei, Graduate Student - Université Paris Cité.
- Samuel Murail, Associate Professor - Université Paris Cité, CMPLI, RPBS platform.
See also the list of contributors who participated in this project.
Citing this work
If you use the code of this package, please cite:
- Reguei A and Murail S. Af-analysis: a Python package for Alphafold analysis.
Journal of Open Source Software (2025) doi: 10.21105/joss.07577
bibtex
@Article{reguei_af-analysis_2025,
title = {Af-analysis: a {Python} package for {Alphafold} analysis},
volume = {10},
issn = {2475-9066},
shorttitle = {Af-analysis},
url = {https://joss.theoj.org/papers/10.21105/joss.07577},
doi = {10.21105/joss.07577},
language = {en},
number = {107},
urldate = {2025-03-14},
journal = {Journal of Open Source Software},
author = {Reguei, Alaa and Murail, Samuel},
month = mar,
year = {2025},
pages = {7577},
}
License
This project is licensed under the GNU General Public License version 2 - see the LICENSE file for details.
References
- Jumper et al. Nature (2021) doi: 10.1038/s41586-021-03819-2
- Abramson et al. Nature (2024) doi: 10.1038/s41586-024-07487-w
- Mirdita et al. Nature Methods (2022) doi: 10.1038/s41592-022-01488-1
- Evans et al. bioRxiv (2021) doi: 10.1101/2021.10.04.463034
- Bryant et al. Nat. Commun. (2022) doi: 10.1038/s41467-022-28865-w
- Zhu et al. Bioinformatics (2023) doi: 10.1093/bioinformatics/btad424
- Kim et al. bioRxiv (2024) doi: 10.1101/2024.02.19.580970
- Yu et al. Bioinformatics (2023) doi: 10.1093/bioinformatics/btac749
- Wohlwend et al. bioRxiv (2024) doi: 10.1101/2024.11.19.624167
- Chai Discovery et al. bioRxiv (2024) doi:10.1101/2024.10.10.615955v2
- MassiveFold Raouraoua et al. Nat. Comput. Sci. (2024) doi:10.1038/s43588-024-00714-4
Owner
- Name: Samuel Murail
- Login: samuelmurail
- Kind: user
- Location: Paris
- Company: Université de Paris
- Website: https://samuelmurail.github.io/PersonalPage/
- Repositories: 6
- Profile: https://github.com/samuelmurail
Assistant Professor of bioinformatics
JOSS Publication
Af-analysis: a Python package for Alphafold analysis
Authors
Tags
Alphafold Protein Structure Structural bioinformaticsCitation (CITATION.cff)
cff-version: "1.2.0"
authors:
- family-names: Reguei
given-names: Alaa
orcid: "https://orcid.org/0009-0006-1718-4864"
- family-names: Murail
given-names: Samuel
orcid: "https://orcid.org/0000-0002-3842-5333"
contact:
- family-names: Murail
given-names: Samuel
orcid: "https://orcid.org/0000-0002-3842-5333"
doi: 10.5281/zenodo.14859764
message: If you use this software, please cite our article in the
Journal of Open Source Software.
preferred-citation:
authors:
- family-names: Reguei
given-names: Alaa
orcid: "https://orcid.org/0009-0006-1718-4864"
- family-names: Murail
given-names: Samuel
orcid: "https://orcid.org/0000-0002-3842-5333"
date-published: 2025-03-14
doi: 10.21105/joss.07577
issn: 2475-9066
issue: 107
journal: Journal of Open Source Software
publisher:
name: Open Journals
start: 7577
title: "Af-analysis: a Python package for Alphafold analysis"
type: article
url: "https://joss.theoj.org/papers/10.21105/joss.07577"
volume: 10
title: "Af-analysis: a Python package for Alphafold analysis"
GitHub Events
Total
- Create event: 3
- Issues event: 5
- Release event: 2
- Watch event: 16
- Issue comment event: 9
- Push event: 53
- Fork event: 3
Last Year
- Create event: 3
- Issues event: 5
- Release event: 2
- Watch event: 16
- Issue comment event: 9
- Push event: 53
- Fork event: 3
Committers
Last synced: 7 months ago
Top Committers
| Name | Commits | |
|---|---|---|
| samuelmurail | s****l@u****r | 198 |
| Alaa REGUEI | 1****a | 2 |
Committer Domains (Top 20 + Academic)
Issues and Pull Requests
Last synced: 6 months ago
All Time
- Total issues: 6
- Total pull requests: 2
- Average time to close issues: 20 days
- Average time to close pull requests: 1 day
- Total issue authors: 4
- Total pull request authors: 1
- Average comments per issue: 0.5
- Average comments per pull request: 0.0
- Merged pull requests: 2
- Bot issues: 0
- Bot pull requests: 0
Past Year
- Issues: 4
- Pull requests: 0
- Average time to close issues: about 3 hours
- Average time to close pull requests: N/A
- Issue authors: 4
- Pull request authors: 0
- Average comments per issue: 0.0
- Average comments per pull request: 0
- Merged pull requests: 0
- Bot issues: 0
- Bot pull requests: 0
Top Authors
Issue Authors
- tubiana (3)
- KuangHu (1)
- BuseSahin1 (1)
- srmnitc (1)
Pull Request Authors
- regueiala (2)
Top Labels
Issue Labels
Pull Request Labels
Packages
- Total packages: 1
-
Total downloads:
- pypi 77 last-month
- Total dependent packages: 0
- Total dependent repositories: 0
- Total versions: 3
- Total maintainers: 1
pypi.org: af-analysis
`AF analysis` is a python library allowing analysis of Alphafold results.
- Homepage: https://github.com/samuelmurail/af_analysis
- Documentation: https://af-analysis.readthedocs.io/
- License: GPL-2.0
-
Latest release: 0.1.4
published about 1 year ago



