bibtexautocomplete
Python package to autocomplete bibtex bibliographies
Science Score: 77.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
✓CITATION.cff file
Found CITATION.cff file -
✓codemeta.json file
Found codemeta.json file -
✓.zenodo.json file
Found .zenodo.json file -
✓DOI references
Found 3 DOI reference(s) in README -
✓Academic publication links
Links to: arxiv.org, zenodo.org -
✓Committers with academic emails
1 of 3 committers (33.3%) from academic institutions -
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (12.1%) to scientific vocabulary
Keywords
Repository
Python package to autocomplete bibtex bibliographies
Basic Info
Statistics
- Stars: 101
- Watchers: 3
- Forks: 6
- Open Issues: 2
- Releases: 8
Topics
Metadata Files
README.md
Bibtex Autocomplete
bibtex-autocomplete or btac is a simple script to autocomplete BibTeX bibliographies. It reads a BibTeX file and looks online for any additional data to add to each entry. It can quickly generate entries from minimal data (a lone title is often sufficient to generate a full entry). You can also use it to only add specific fields (like DOIs, or ISSN) to a manually curated bib file.
It is designed to be as simple to use as possible: just give it a bib file and let btac work its magic! It combines multiple sources and runs consistency and normalization checks on the added fields (only adds URLs that lead to a valid webpage, DOIs that exist at https://dx.doi.org/, ISSN/ISBN with valid check digits...).
It attempts to complete a BibTeX file by querying the following domains: - openalex.org: ~240 million entries - www.crossref.org: ~150 million entries - arxiv.org: open access archive, ~2.4 million entries - semanticscholar.org: ~215 million entries - unpaywall.org: database of open access articles, ~48 million entries - dblp.org: computer science, ~7 million entries - researchr.org: computer science - inspirehep.net: high-energy physics, ~1.5 million entries
Big thanks to all of them for allowing open, easy and well-documented access to their databases. This project wouldn't be possible without them. You can easily narrow down the list of sources if some aren't relevant using command line options.
Contents
- Demo
- Quick overview
- Installation
- Usage
- Command line arguments
- Running from python
- Credit and license
Demo
Quick overview
How does it find matches?
btac queries the websites using the entry DOI (if known) or its title. So
entries that don't have one of those two fields will not be completed.
- DOIs are only used if they can be recognized, so the doi field should
contain "10.xxxx/yyyy" or an URL ending with it.
- Titles should be the full title. They are compared excluding case and
punctuation, but titles with missing words will not match.
- If one or more authors are present, entries with no common authors will not
match. Authors are compared using lower case last names only. Be sure to use
one of the correct BibTeX formats for the author field:
bibtex
author = {First Last and Last, First and First von Last}
(see
https://www.bibtex.com/f/author-field/
for full details)
- If the year is known, entries with different years will also not match.
Disclaimers:
There is no guarantee that the script will find matches for your entries, or that the websites will have any data to add to your entries, (or even that the website data is correct).
The script is designed to minimize the chance of false positives - that is adding data from another similar-ish entry to your entry. If you find any such false positive please report them using the issue tracker.
How are entries completed?
Once responses from all websites have been found, the script will add fields
by performing a majority vote among the sources. To do so it uses smart
normalization and merging tactics for each field:
- Authors (and editors) match if they have same last names and, if both first
names present, the first name of one is equal/an abbreviation of the other.
Author lists match they have at least one author in common.
- ISSN and ISBN are normalized and have their check digits verified. ISBN are converted
to their 13 digit representation.
- URL and DOI are checked for valid format, and further validated by querying
them online to ensure they exist. DOI are normalized to strip any leading URL
and converted to lowercase.
- Many fields match with abbreviation detection (journal, institution, booktitle,
organization, publisher, school and series). So ACM will match
Association for Computer Machinery.
- Pages are normalized to use -- as separator.
- All other fields are compared excluding case and punctuation.
The script will not overwrite any user given non-empty fields, unless the
-f/--force-overwrite flag is given. If you want to check what fields are
added, you can use -v/--verbose to have them printed to standard output (with
source information), or -p/--prefix to have the new fields be prefixed with
BTAC in the output file.
Installation
Can be installed with pip :
console
pip install bibtexautocomplete
You should now be able to run the script using either command:
console
btac --version
python3 -m bibtexautocomplete --version
Note: pip no longer allows installing scripts globally in systems with other
package managers (like most Linux distros). You can install the script locally in
a virtual environment or globally
using pipx:
console
sudo apt install pipx
pipx install bibtexautocomplete
Shell tab completion
If you want tab based completion for btac in your shell, you must install
the optional argcomplete dependency.
```bash
Either install the package separately
pip install argcomplete
Or as a btac optional dependency
pip install bibtexautocomplete[tab]
You then need to register the tab auto-completer. On bash/zsh:
- You can activate completion just for this script with
bash
eval "$(register-python-argcomplete btac)"
For repeated use, I recommend adding this line to your `.bashrc` or `.bash_profile`.
- Alternatively, you can activate completion for all python scripts
using argcomplete by running
bash
activate-global-python-argcomplete
```
and then restarting your shell
If using another shell than bash/zsh on Linux or MacOS, support is not guaranteed. See github.com/kislyuk/argcomplete/contrib for instructions on getting it working on other shells.
Dependencies
This package has two dependencies (automatically installed by pip) :
- bibtexparser (<2.0.0)
- alive_progress (>= 3.0.0) for the fancy progress bar
It also has an optional dependency, argcomplete for tab based completion. It is installed if you pip install bibtexautocomplete[tab].
Usage
The command line tool can be used as follows:
console
btac [--flags] <input_files>
Examples :
btac my/db.bib: reads from./my/db.bib, writes to./my/db.btac.bib. A different output file can be specified with-o.btac -i db.bib: reads fromdb.biband overwrites it (inplace flag). Avoid on non backed-up/version-controlled files, I'd hate it if my script corrupted your data.btac folder: reads from all files ending with.bibin folder. Excludes.btac.bibfiles unless they are the only.bibfiles present. Writes tofolder/file.btac.bibunless inplace flag is set.btacwith no inputs is same asbtac ., reads file from current working directorybtac -c doi ...only completes DOI fields, leave others unchangedbtac -v ...verbose mode, pretty prints all new fields when done. See this image for a preview of verbose output.
Note: the parser doesn't preserve format information, so this script will reformat your files. Some formatting options are provided to control output format.
Slow responses: Sometimes due to server traffic, a source DB may take significantly longer
to respond and slow btac.
- You can increase timeout with btac ... -t 60 (60s) or btac ... -t -1 (no timeout)
- You can disable queries to the offender btac ... -Q <website>
- You can try again at another time
Command line arguments
As btac has a lot of option I'd recommend setting up an alias if you use a lot
regularly.
Specifying output
-o --output <file.bib>
Write output to given file. Can be used multiple times when also giving
multiple inputs. Maps inputs to outputs in order. If there are extra inputs,
uses default name (old_name.btac.bib). Ignored in inplace (-i) mode.
For example btac db1.bib db2.bib db3.bib -o out1.bib -o out2.bib reads db1.bib,
db2.bib and db3.bib, and write their outputs to out1.bib, out2.bib
and db3.btac.bib respectively.
-i --inplaceModify input files inplace, ignores any specified output files. Avoid on non backed-up/version-controlled files, I'd hate it if my script corrupted your data.-O --no-outputdon't write any output files (except the one specified by--dump-data) can be used with-v/--verbosemode to only print a list of changes to the terminal
Query filtering
-q --only-query <site>or-Q --dont-query <site>
Restrict which websites to query from. <site> must be one of: openalex,
crossref, arxiv, s2, unpaywall, dblp, researchr, hep. These arguments
can be used multiple times, for example to only query Crossref and DBLP use
-q crossref -q dblp or
-Q openalex -Q researchr -Q unpaywall -Q arxiv -Q s2 -Q hep
-e --only-entry <id>or-E --exclude-entry <id>
Restrict which entries should be autocompleted. <id> is the entry ID used in
your BibTeX file (e.g. @inproceedings{<id> ... }). These arguments can also
be used multiple times to select only/exclude multiple entries
--sf --start-from <id>
Only complete the entries that come after the given id (inclusive). This is useful when resuming a previously interrupted auto-completion on the same file.
-c --only-complete <field>or-C --dont-complete <field>
Restrict which fields you wish to autocomplete. Field is a BibTeX field (e.g.
author, doi,...). So if you only wish to add missing DOIs use -c doi.
-b --filter-fields-by-entrytype <required|optional|all>only add fields that correspond to the given entry type in bibtex's data model. Disabled by default.requiredonly adds required fields,optionaladds required and optional fields, andalladds required, optional and non-standard fields (doi, issn and isbn). A list of required/optional fields by entry type can be found on the tex stackexchange-w --overwrite <field>or-W --dont-overwrite <field>
Force overwriting of the selected fields. If using -W author -W journal
your force overwrite of all fields except author and journal. The
default is to override nothing (only complete absent and blank fields).
For a more complex example btac -C doi -w author means complete all fields
save DOI, and only overwrite author fields.
You can also use the -f flag to overwrite everything or the -p flag to add
a prefix to new fields, thus avoiding overwrites.
-m --markand-M --ignore-mark
This is useful to avoid repeated queries if you want to run btac many times
on the same (large) file.
By default, btac ignores any entry with a BTACqueried field. --ignore-mark
overrides this behavior.
When --mark is set, btac adds a BTACqueried = {yyyy-mm-dd} field to each entry
it queries.
New field formatting
You can use the following arguments to control how btac formats the new fields
- --fu --escape-unicode replace unicode symbols by latex escapes sequence (for
example: replace é with {\'e}). The default is to keep unicode symbols as is.
- --fp --protect-uppercase <field> or --FP --dont-protect-uppercase <field> or
--fpa --protect-all-uppercase, insert braces around words containing uppercase
letters in the given fields to ensure bibtex will preserve them. The three
arguments are mutually exclusive, and the first two can be used multiple times
to select/deselect multiple fields.
Global output formatting
Unfortunately bibtexparser doesn't preserve format information, so this script will reformat your BibTeX file. Here are a few options you can use to control the output format:
--fa --align-valuespad field names to align all values
bibtex
@article{Example,
author = {Someone},
doi = {10.xxxx/yyyyy},
}
--fc --comma-firstuse comma first syntax
bibtex
@article{Example
, author = {Someone}
, doi = {10.xxxx/yyyyy}
,
}
--fl --no-trailing-commadon't add the last trailing comma--fi --indent <space>space used for indentation, default is a tab. Can be specified as a number (number of spaces) or a string with spaces and_,t, andncharacters to mark space, tabs and newlines.
Optional flags
-p --prefixWrite new fields with a prefix. The script will addBTACtitle = ...instead oftitle = ...in the bib file. This can be combined with-fto safely show info for already present fields.
Note that this can overwrite existing fields starting with BTACxxxx, even
without the -f option.
- -f --force-overwrite Overwrite already present fields. The default is to
overwrite a field only if it is empty or absent
- -D --diff only print the new fields in the output file. In this mode, old
fields are removed and entries with no new fields are deleted. This cannot be
used with the -i --inplace flag for safety reasons. If you really want to overwrite
your input file (and delete a bunch of data in the process), you can do so with
by specifying it explicitly via the -o --output option.
- -u --copy-doi-to-url If a DOI is found but no URL, set the URL field
to https://dx.doi.org/<doi>
-t --timeout <float>set timeout on request in seconds, default: 20.0 s, increase this if you are getting a lot of timeouts. Set it to -1 for no timeout.-S --ignore-sslbypass SSL verification. Use this if you encounter the error:[SSL: CERTIFICATE_VERIFY_FAILED] certificate verify failed: certificate has expired (_ssl.c:1129)Another (better) fix for this is to runpip install --upgrade certifito update python's certificates.--ns --no-skipdisable skipping. By default, btac will skip queries to sources if they lag behind (>=10 queries remain or >=60s delay between queries) when 2/3rds of the other sources have completed. This avoids having a single source slow down btac considerably.-d --dump-data <file.json>writes matching entries to the given JSON files.
This allows to see duplicate fields from different sources that are otherwise overwritten when merged into a single entry.
The JSON file will have the following structure:
json
[
{
"entry": "<entry_id>",
"new-fields": 8,
"crossref": {
"query-url": "https://api.crossref.org/...",
"query-response-time": 0.556,
"query-response-status": 200,
"author" : "Lastname, Firstnames and Lastname, Firstnames ...",
"title" : "super interesting article!",
"..." : "..."
},
"openalex": ...,
"arxiv": null, // null when no match found
"unpaywall": ...,
"dblp": ...,
"researchr": ...,
"inspire": ...
},
...
]
-v --verboseverbose mode shows more info. It details entries as they are being processed and shows a summary of new fields and their source at the end. Using it more than once prints debug info (up to four times).
Verbose mode looks like this:
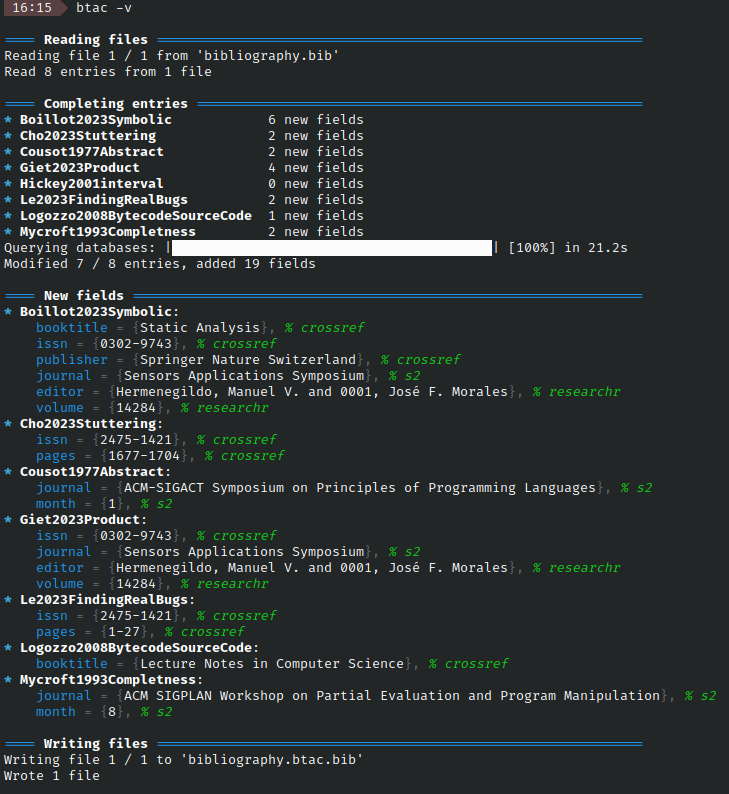 -
- -s --silent hide info and progress bar. Keep showing warnings and errors.
Use twice to also hide warnings, thrice to also hide errors and four times to
also hide critical errors, effectively killing all output.
- --color <auto|always|never> sets whether btac should use colored output.
Can also be set by the NO_COLOR or the CLICOLOR_FORCE environment variables,
as explained here: http://bixense.com/clicolors/. Defaults to auto, which checks
if standard output is a terminal via the isatty function
--versionshow version number-h --helpshow help
Running from python
You can call the main function of this script from python directly, specifying a list of arguments as you would on the command line: ```python from bibtexautocomplete import main
main(["file.bib", "-o", "output.bib"]) ```
For more interactivity and more varied inputs/outputs than reading from files and
writing to files, use the BibtexAutocomplete class. Here is a small demonstration:
```python
from bibtexautocomplete import BibtexAutocomplete
1 - Create a BibtexAutocomplete instance with the desired settings
Note: some settings are stored in global variables, so avoid having multiple
instances of this class in parrallel
completer = BibtexAutocomplete(**settings)
2 - Load a Bibtex information using any of the following
2.1 - Load a single file or list of files
completer.loadfile("ex.bib") completer.loadfile(["ex1.bib", "ex2.bib"])
2.2 - Load bibtex content as string or list of strings
completer.loadstring(bibtex) completer.loadstring([bibtex1, bibtex2])
2.3 - Load entry, list of entries, or list of list of entries as a dict
Lowercase name for fields, "ID" and "ENTRYTYPE" for entry id and type
completer.load_entry({ "author": "John Doe", "title": "My Awesome Paper", "ID": "foo", "ENTRYTYPE": "article" })
You can also specify author and editor fields as a (list of) firstnames and lastnames
completer.load_entry({ "author": [{"firstname": "John"}, {"lastname": "Doe"}], "title": "My Awesome Paper", "ID": "foo", "ENTRYTYPE": "article" })
3 - Run the completer (may take a while)
completer.autocomplete()
4 - Get the results
As the input may be split into mutliple files (or strings), so is the output
The length of output list is the sum of:
- the length of lists passed to loadfile and loadstring
- the number of calls to load_entry
When using write_file, you must provide the same number of filepaths
completer.writefile("ex.btac.bib") completer.writefile(["ex1.bib", "ex2.bib"]) completer.writestring() # type: list[str] completer.writeentry() # type: list[list[EntryType]] ```
The settings passed to the BibtexAutocomplete constructor mirror the
command-line arguments, see their documentation for
details.
```python
lookups: Iterable[LookupType] = ...,
Specify which entries should be completed (default: all)
entries: Optional[Container[str]] = None, mark: bool = False, ignoremark: bool = False, prefix: bool = False, escapeunicode: bool = False, diff_mode: bool = False,
Restrict which fields should be completed (default: all)
fieldstocomplete: Optional[Set[FieldType]] = None,
Specify which fields should be overwritten (default: none)
fieldstooverwrite: Optional[Set[FieldType]] = None,
Specify which fields should have uppercase protection (default: none)
fieldstoprotectuppercase: Container[str] = set(), filterbyentrytype: Literal["no", "required", "optional", "all"] = "no", copydoitourl: bool = False, startfrom: Optional[str] = None, dontskipslowqueries: bool = False, timeout: Optional[float] = 20, # Timeout on all queries, in seconds ignore_ssl: bool = False, # Bypass SSL verification verbose: int = 0, # Verbosity level, from 4 (very verbose debug) to -3 (no output)
Output formatting
alignvalues: bool = False, commafirst: bool = False, notrailingcomma: bool = False, indent: str = "\t", ```
Credit and license
This project was first inspired by the solution provided by thando in this TeX stack exchange post. I worked on as part of a course on Web data management in 2021-2022 as part of my masters (MPRI).
This project is free and open-source. It is distributed under terms of the MIT License. See the LICENSE file for more information
Owner
- Name: Dorian Lesbre
- Login: dlesbre
- Kind: user
- Location: Paris, France
- Website: https://www.eleves.ens.fr/home/dlesbre
- Repositories: 18
- Profile: https://github.com/dlesbre
Master's student in computer science. École Normale Supérieure Paris
Citation (CITATION.cff)
cff-version: 1.2.0
message: "If you use this software, please cite it as below."
authors:
- family-names: "Lesbre"
given-names: "Dorian"
orcid: "https://orcid.org/0000-0002-4328-6753"
title: "Bibtex autocomplete"
version: 1.4.0
doi: 10.5281/zenodo.13998520
date-released: 2024-10-27
url: "https://github.com/dlesbre/bibtex-autocomplete"
GitHub Events
Total
- Create event: 6
- Issues event: 3
- Release event: 3
- Watch event: 12
- Delete event: 4
- Issue comment event: 3
- Push event: 26
Last Year
- Create event: 6
- Issues event: 3
- Release event: 3
- Watch event: 12
- Delete event: 4
- Issue comment event: 3
- Push event: 26
Committers
Last synced: almost 3 years ago
All Time
- Total Commits: 242
- Total Committers: 3
- Avg Commits per committer: 80.667
- Development Distribution Score (DDS): 0.041
Top Committers
| Name | Commits | |
|---|---|---|
| Dorian Lesbre | d****e@g****m | 232 |
| Dorian Lesbre | 5****e@u****m | 9 |
| Dorian Lesbre | d****e@e****r | 1 |
Committer Domains (Top 20 + Academic)
Issues and Pull Requests
Last synced: 6 months ago
All Time
- Total issues: 18
- Total pull requests: 0
- Average time to close issues: 24 days
- Average time to close pull requests: N/A
- Total issue authors: 12
- Total pull request authors: 0
- Average comments per issue: 2.33
- Average comments per pull request: 0
- Merged pull requests: 0
- Bot issues: 0
- Bot pull requests: 0
Past Year
- Issues: 5
- Pull requests: 0
- Average time to close issues: 2 months
- Average time to close pull requests: N/A
- Issue authors: 3
- Pull request authors: 0
- Average comments per issue: 3.2
- Average comments per pull request: 0
- Merged pull requests: 0
- Bot issues: 0
- Bot pull requests: 0
Top Authors
Issue Authors
- tomkoenecke (3)
- dlesbre (3)
- homocomputeris (2)
- edzob (2)
- LukasCBossert (1)
- pixelcraftAU (1)
- leonmoonen (1)
- yipeah (1)
- daniellobrunello (1)
- KonradHoeffner (1)
- matee911 (1)
- 00sapo (1)
- schulzch (1)
Pull Request Authors
Top Labels
Issue Labels
Pull Request Labels
Packages
- Total packages: 1
-
Total downloads:
- pypi 134 last-month
- Total dependent packages: 0
- Total dependent repositories: 1
- Total versions: 28
- Total maintainers: 1
pypi.org: bibtexautocomplete
Script to autocomplete bibtex files by polling online databases
- Homepage: https://github.com/dlesbre/bibtex-autocomplete
- Documentation: https://bibtexautocomplete.readthedocs.io/
- License: MIT License Copyright (c) 2022-2025 Dorian Lesbre Permission is hereby granted, free of charge, to any person obtaining a copy of this software and associated documentation files (the "Software"), to deal in the Software without restriction, including without limitation the rights to use, copy, modify, merge, publish, distribute, sublicense, and/or sell copies of the Software, and to permit persons to whom the Software is furnished to do so, subject to the following conditions: The above copyright notice and this permission notice shall be included in all copies or substantial portions of the Software. THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.
-
Latest release: 1.4.3
published 6 months ago
Rankings
Maintainers (1)
Dependencies
- actions/checkout v3 composite
- actions/setup-python v3 composite
- python 3.8-slim build
- alive-progress >=3.0.0
- bibtexparser <2.0.0









