Recent Releases of cdk
cdk - CDK 2.10
New Features/Key Changes
AtomContainer new implementation (IMPORTANT)
The new AtomContainer implementation is now the default after a gradual introduction. You can still use the old implementation but you must explicitly create an AtomContainerLegacy. This should be a seamless change for most but please notify if you have an unexpected error.
SMIRKS
SMIRKS support with the ability to approximate other implementations (inc. Daylight and RDKit Reaction Smarts). It includes convenience APIs for applying a transform to all places at once (i.e. dt_xapply) and efficient support for hydrogen handling (explicit hydrogen are not required on the input). Overall the speed it good and a transform can be run over all of ChEMBL 35 in only ~30 seconds (see Appendix A1).
```java IChemObjectBuilder bldr = SilentChemObjectBuilder.getInstance(); SmilesParser smipar = new SmilesParser(bldr); SmilesGenerator smigen = new SmilesGenerator(SmiFlavor.Default);
String smminp = "c1cc(N(=O)=O)ccc1N(=O)=O"; IAtomContainer mol = smipar.parseSmiles(smminp); Smirks.compile("N:1=[OD1+0]>>N+:1[O-] polar-nitro") .apply(mol); // exclusive apply mode String smiout = smigen.create(mol); // C1=CC(N+[O-])=CC=C1N+[O-] ```
More information can be found in the JavaDoc and functionality will be added to the CDK Depict Web Application.
Reaction InChI (RInChI) generation
A pure Java implementation of Reaction InChI has been added allowing generation of RInChI strings and keys:
java
IChemObjectBuilder bldr = SilentChemObjectBuilder.getInstance();
SmilesParser smipar = new SmilesParser(bldr);
IReaction reaction = smipar.parseReactionSmiles("CCO.[CH3:1][C:2](=[O:3])[OH:4]>[H+]>CC[O:4][C:2](=[O:3])[CH3:1].O Ethyl esterification [1.7.3]\n");
RInChIGenerator rinchigen = new RInChIGenerator();
rinchigen.generate(reaction);
System.err.println(rinchigen.getRInChI()); // RInChI=1.00.1S/C2H4O2/c1-2(3)4/h1H3,(H,3,4)!C2H6O/c1-2-3/h3H,2H2,1H3<>C4H8O2/c1-3-6-4(2)5/h3H2,1-2H3!H2O/h1H2<>p+1/d+
System.err.println(rinchigen.getShortRInChIKey()); // Short-RInChIKey=SA-FUHFF-JJFIATRHOH-UDXZTNISGZ-GPRLSGONYQ-NUHFF-NUHFF-NUHFF-ZZZ
RDfile reading support
RDfiles belong to the CT file family formats and allows records with associated experimental data.
java
RdfileReader rdReader = new RdfileReader(new FileReader("/tmp/pistachio-rxns-2501091627.rd"),
SilentChemObjectBuilder.getInstance(),
true);
while (rdReader.hasNext()) {
RdfileRecord record = rdReader.next();
if (record.isRxnFile()) {
IReaction reaction = record.getReaction();
} else {
IAtomContainer container = record.getAtomContainer();
}
}
Faster ring and aromaticity perception
Faster ring membership and aromaticity assignment. The move to AtomContainer2 (see above) allows additional optimizations to these algorithms. The APIs will run faster however for aromaticity you must use Cycles.all() on it's own. There is also a new static method for convenience and improved aromatic model encoding.
```java // new way, no checked exception Cycles.markRingAtomsAndBonds(molecule); // prerequisite if (!Aromaticity.apply(Aromaticity.Model.Daylight, molecule)) { // return false = too many cycles to check }
// old way (will still be faster) Aromaticity aromaticity = new Aromaticity(ElectronDonation.daylight(), Cycles.all()); IAtomContainer container = ...; try { if (aromaticity.apply(molecule)) { // } } catch (CDKException e) { // cycle computation was intractable } ```
Improved inorganic stereochemistry
It is now possible to represent degenerate inorganic stereochemistry where one or more neighbours are missing/implicit. For example, we can describe a square pyramidal structure as an octahedral without a missing ligand. Support for implicit/explicit hydrogens around theses atoms has also been improved.
[NH3][Co@OH25](Cl)(Cl)(Cl)(Cl) sqpyr
[NH3][Co@OH4](Cl)(Cl)[NH3] seesaw
You can also use this in SMARTS to match across atoms and equatorial using the following patterns:
Cl[Co@OH1]Cl across
Cl[Co@OH3]Cl equatorial
Functional Group Finder
A functional group finder has been added based on Peter Ertl's algorithm.
Peter Ertl. 2017 Fritsch et al. 2019
The API allows you generate the functional groups as fragments or my favorite which is fill an array with identifier numbers - this is then very easy to depict.
```java IChemObjectBuilder bldr = SilentChemObjectBuilder.getInstance(); SmilesParser smipar = new SmilesParser(bldr);
String smiles = "C2C(NC)=NC3=C(C(C1=CC=CC=C1)=N2=O)C=C(Cl)C=C3"; IAtomContainer mol = smipar.parseSmiles(smiles); FunctionalGroupsFinder fgFinder = FunctionalGroupsFinder.withNoEnvironment();
Cycles.markRingAtomsAndBonds(mol); Aromaticity.apply(Aromaticity.Model.Daylight, mol);
// extract the groups as new fragments
List
// fill an array with numbers that indicate which functional group something belongs to int[] fgrps = new int[mol.getAtomCount()]; fgFinder.find(fgrps, mol);
// Set the group as the atom map/class in SMILES
for (IAtom atom : mol.atoms())
atom.setMapIdx(1+fgrps[atom.getIndex()]);
System.out.println(new SmilesGenerator(SmiFlavor.AtomAtomMap).create(mol));
```
Sugar Moiety Removal
The Sugar Removal Utility (SRU) implements a generalized algorithm for automated detection of circular and linear sugars in molecular structures and their removal.
Convenience APIs
- Iterate over molecules of a reaction and sets
- Creating atoms/bonds in the context of molecules with:
mol.newAtom()andmol.newBond()and others. - Better IO error handling
Contributors
55 Egon Willighagen
8 Felix Bänsch
3 Jean Marois
245 John Mayfield
43 Jonas Schaub
2 Matthias Mailänder
5 Tyler Peryea
123 Uli Fechner
3 Valentyn Kolesnikov
3 Stefan Kuhn
Overview of Pull Requests
- SonarCloud is not reporting test coverage correctly because it was no… by @johnmay in https://github.com/cdk/cdk/pull/1000
- Improved the abbreviation handling over atom sets, this is useful for… by @johnmay in https://github.com/cdk/cdk/pull/996
- Fix - avoid placing a wedge on the right-angled bond when a centre is… by @johnmay in https://github.com/cdk/cdk/pull/998
- Quality of life API interfaces. The IAtomContainerSet and IReaction c… by @johnmay in https://github.com/cdk/cdk/pull/997
- Sonar settings for aggregated test coverage. by @johnmay in https://github.com/cdk/cdk/pull/1001
- CMLXOM 4.6 by @egonw in https://github.com/cdk/cdk/pull/1004
- Redo @parit's changes for net/undirected reaction depiction on the ne… by @johnmay in https://github.com/cdk/cdk/pull/1009
- Smiles 0 isotope by @johnmay in https://github.com/cdk/cdk/pull/1007
- When atoms/bonds are aware of the container they are in - it is usefu… by @johnmay in https://github.com/cdk/cdk/pull/1010
- Fix the CDK C.plus atom type, there was already comment in the test t… by @johnmay in https://github.com/cdk/cdk/pull/1011
- Query bond funcs by @johnmay in https://github.com/cdk/cdk/pull/938
- Read the atom-atom mapping info from a V3000 file. by @johnmay in https://github.com/cdk/cdk/pull/1012
- Fixes https://github.com/cdk/depict/issues/76. We do not like -C=CO a… by @johnmay in https://github.com/cdk/cdk/pull/1015
- Added an API for fatal IO errors by @egonw in https://github.com/cdk/cdk/pull/1019
- Java21 by @johnmay in https://github.com/cdk/cdk/pull/1014
- Fixes #1024 - we should perhaps rework the CDK radical representation… by @johnmay in https://github.com/cdk/cdk/pull/1025
- Updated dependencies by @egonw in https://github.com/cdk/cdk/pull/1026
- Fix for non-deterministic CIP designation bug by @tylerperyea in https://github.com/cdk/cdk/pull/1027
- Fix and issue with contraction on terminal attachment points. by @johnmay in https://github.com/cdk/cdk/pull/1028
- Fix a minor issue with an abbreviation like -NnButBu. Currently this … by @johnmay in https://github.com/cdk/cdk/pull/1030
- Make sure Sgroups attached to reactions get passed through and emitte… by @johnmay in https://github.com/cdk/cdk/pull/1031
- First pass at aligned depictions API. by @johnmay in https://github.com/cdk/cdk/pull/1032
- Symmetry calculation may fail. by @johnmay in https://github.com/cdk/cdk/pull/1033
- Code cleanup by @egonw in https://github.com/cdk/cdk/pull/1018
- Fix a minor issue from sonarcloud, we check the counts elsewhere so t… by @johnmay in https://github.com/cdk/cdk/pull/1034
- Additional tokens reagent label formatting. by @johnmay in https://github.com/cdk/cdk/pull/1036
- Depict align tweaks by @johnmay in https://github.com/cdk/cdk/pull/1037
- Added a missing test class for Elements by @egonw in https://github.com/cdk/cdk/pull/1042
- Added isMetalloid utility method to Elements class by @JonasSchaub in https://github.com/cdk/cdk/pull/1041
- Refine OSGi import rules by @Mailaender in https://github.com/cdk/cdk/pull/1043
- AtomContainer2 Phase 2 by @johnmay in https://github.com/cdk/cdk/pull/1047
- New convenience methods on the Atom API. by @johnmay in https://github.com/cdk/cdk/pull/1046
- AtomContainer2 phase 3 by @johnmay in https://github.com/cdk/cdk/pull/1048
- Binconnected - faster ring atom/bond marking by @johnmay in https://github.com/cdk/cdk/pull/1051
- Add transform/SMIRKS support to CDK. by @johnmay in https://github.com/cdk/cdk/pull/916
- The number of essential/relevant cycles can be exponential for some m… by @johnmay in https://github.com/cdk/cdk/pull/1052
- Relavent cycles limit test by @johnmay in https://github.com/cdk/cdk/pull/1053
- Updated JNA-InChI (JNA compatibility) by @egonw in https://github.com/cdk/cdk/pull/1054
- Create CITATION.cff by @egonw in https://github.com/cdk/cdk/pull/1055
- Fix a corner case when depicting cc(C)c by @johnmay in https://github.com/cdk/cdk/pull/1059
- small doc fix for cdkAllowingExocyclic() by @JonasSchaub in https://github.com/cdk/cdk/pull/1060
- Increase the max fragment count when generating abbreviations. by @johnmay in https://github.com/cdk/cdk/pull/1066
- Ensure the ESSSR parameter is reported in the FP version info. by @johnmay in https://github.com/cdk/cdk/pull/1065
- Updated ${version} in pom.xml by @javadev in https://github.com/cdk/cdk/pull/1070
- CMLXOM 4.9 and log4j 2.23.1 by @egonw in https://github.com/cdk/cdk/pull/1072
- Link to the ChemPyFormatics 'book' by @egonw in https://github.com/cdk/cdk/pull/1071
- Maven build system updates by @egonw in https://github.com/cdk/cdk/pull/1074
- Only run JaCoCo once by @egonw in https://github.com/cdk/cdk/pull/1076
- Depiction issues by @johnmay in https://github.com/cdk/cdk/pull/1080
- Moved a number of test classes to the same module as the tested classes by @egonw in https://github.com/cdk/cdk/pull/1081
- Only copy mapped bonds when deciding how to align the structure. by @johnmay in https://github.com/cdk/cdk/pull/1082
- Fix a bug in the MDLV2000Reader where the wrong "molecule" is used. by @johnmay in https://github.com/cdk/cdk/pull/1085
- Removed a module that has been empty for a few years by @egonw in https://github.com/cdk/cdk/pull/1087
- The path based fingerprint should be identical with/without explicit … by @johnmay in https://github.com/cdk/cdk/pull/1089
- Stabilise the CDK atom type based aromaticity model. This causes a sm… by @johnmay in https://github.com/cdk/cdk/pull/1091
- Use interfaces instead of instances and use silent by @egonw in https://github.com/cdk/cdk/pull/1094
- Recovers the "simple" patches from testing2 by @egonw in https://github.com/cdk/cdk/pull/1093
- Improving the testing coverage by @egonw in https://github.com/cdk/cdk/pull/1095
- Overhaul and optimise the aromaticity procedures in CDK. by @johnmay in https://github.com/cdk/cdk/pull/1092
- Invalid stereochemistry group causes infinite loop by @marois in https://github.com/cdk/cdk/pull/1098
- add ability to read MDL RXN V3000 files with zero reactants but a REACTANT block by @uli-f in https://github.com/cdk/cdk/pull/1100
- Fix CCD/WebMolKit Sgroups that are missing the SBL. by @johnmay in https://github.com/cdk/cdk/pull/1099
- Fixes code examples by @egonw in https://github.com/cdk/cdk/pull/1104
- support bond type gt4 in MDLV3000Reader by @uli-f in https://github.com/cdk/cdk/pull/1102
- Make sure Atom/Bond's ged deref'd when going into a QueryAtomContainer. by @johnmay in https://github.com/cdk/cdk/pull/1105
- Integration of functional groups identification functionality following the Ertl algorithm by @JonasSchaub in https://github.com/cdk/cdk/pull/1039
- Generally cheminf formats use ASCII and we should not be checking the… by @johnmay in https://github.com/cdk/cdk/pull/1107
- Rdfile reader by @uli-f in https://github.com/cdk/cdk/pull/942
- add javadocs to RdfileReader and RdfileRecord, make RdfileReader final by @uli-f in https://github.com/cdk/cdk/pull/1109
- Checked, updated, and formatted documentation of FunctionalGroupsFinder by @JonasSchaub in https://github.com/cdk/cdk/pull/1110
- added test case in CDKAtomTypeMatcherFilesTest that gives rise to an NPE by @uli-f in https://github.com/cdk/cdk/pull/919
- support bond type gt4 in MDLV3000Writer by @uli-f in https://github.com/cdk/cdk/pull/1106
- Integration of sugar moiety removal functionality by @JonasSchaub in https://github.com/cdk/cdk/pull/1040
- Resolved Sonar issue with addAll in unmodifiable set by @javadev in https://github.com/cdk/cdk/pull/1114
- add test dependencies assertj and mockito-junit-jupiter by @uli-f in https://github.com/cdk/cdk/pull/1115
- add DefaultChemObjectReaderErrorHandler by @uli-f in https://github.com/cdk/cdk/pull/1112
- Fix a bug with the default SMILES output. AtomStereo was not emitted … by @johnmay in https://github.com/cdk/cdk/pull/1116
- Update BEAM to v1.3.7 to fix a corner case with reading SMILES and ar… by @johnmay in https://github.com/cdk/cdk/pull/1117
- Advanced Inorganic Handling by @johnmay in https://github.com/cdk/cdk/pull/1118
- inorganic stereo 2 by @johnmay in https://github.com/cdk/cdk/pull/1120
- Depiction Improvements (Nov 2024) by @johnmay in https://github.com/cdk/cdk/pull/1122
- Fix a minor issue with a NPE on the AwtArea util and improve tests. by @johnmay in https://github.com/cdk/cdk/pull/1123
- Fix (in)saturation expression behaviour by @johnmay in https://github.com/cdk/cdk/pull/1124
- Update CMLCoreModule.java. So far, if there was no order defined for … by @stefhk3 in https://github.com/cdk/cdk/pull/1126
- Patch/stefhk3 patch 2 fix by @johnmay in https://github.com/cdk/cdk/pull/1127
- Create codeql.yml by @javadev in https://github.com/cdk/cdk/pull/1128
- fix crash in SmilesGenerator when calling with reaction not having one or more reaction components by @uli-f in https://github.com/cdk/cdk/pull/1129
- This fixes a problem with the ordering of &. by @stefhk3 in https://github.com/cdk/cdk/pull/1131
- remove deprecated calls from InChIGeneratorTest by @uli-f in https://github.com/cdk/cdk/pull/1134
- add getAgentCount method to IReaction interface by @uli-f in https://github.com/cdk/cdk/pull/1133
- Updated dependencies by @egonw in https://github.com/cdk/cdk/pull/1136
- improve and document agent handling in MDLRXNV2000Reader by @uli-f in https://github.com/cdk/cdk/pull/1138
- update junit-jupiter dependencies to version 5.11.4 by @uli-f in https://github.com/cdk/cdk/pull/1139
- RInChI implementation based on InChI native + Java logic by @uli-f in https://github.com/cdk/cdk/pull/1137
New Contributors
- @tylerperyea made their first contribution in https://github.com/cdk/cdk/pull/1027
- @JonasSchaub made their first contribution in https://github.com/cdk/cdk/pull/1041
- @stefhk3 made their first contribution in https://github.com/cdk/cdk/pull/1126
Full Changelog: https://github.com/cdk/cdk/compare/cdk-2.9...cdk-2.10
Appendix A1
```java IChemObjectBuilder bldr = SilentChemObjectBuilder.getInstance(); SmilesParser smipar = new SmilesParser(bldr); SmilesGenerator smigen = new SmilesGenerator(SmiFlavor.Default);
// -OH => -[O-] SmirksTransform deprotonate = Smirks.compile("[c:1][OX2v2+0:2][H]>>[c:1][O-:2] de-protonate\n");
long tBegin = System.nanoTime(); long tSmirks = 0; int count = 0; try (BufferedReader brdr = new BufferedReader(new FileReader("/data/chembl35.smi")); BufferedWriter bwtr = new BufferedWriter(new FileWriter("/data/chembl35.smi.norm"))) { String line; while ((line = brdr.readLine()) != null) { IAtomContainer mol = smipar.parseSmiles(line); long tSplit0 = System.nanoTime();
// SMIRKS pattern will do aromaticity automatically, if you
// have multiple patterns being applied it may be better
// to turn this of deprotonate.setPrepare(false); and do it
// yourself
boolean changed = deprotonate.apply(mol);
long tSplit1 = System.nanoTime();
tSmirks += (tSplit1-tSplit0);
if (changed)
line = smigen.create(mol) + " " + mol.getTitle();
bwtr.write(line);
bwtr.newLine();
++count;
if (count % 1000 == 0)
System.err.printf("\r%d...", count);
}
} catch (IOException e) { throw new RuntimeException(e); }
long tEnd = System.nanoTime(); long tElapsed = TimeUnit.NANOSECONDS.toMillis(tEnd-tBegin); System.err.printf("\rdone %d in %.3fs (%.0f mol/s)\n", count, tElapsed / 1e3, count / (tElapsed/1e3)); System.err.printf("SMIRKS in %.3fs (%.0f mol/s)\n", tSmirks / 1e9, count / (tSmirks/1e9)); ```
M1 Pro 2021 results:
done 2474590 in 29.449s (84030 mol/s)
SMIRKS in 8.591s (288054 mol/s)
- Java
Published by johnmay 12 months ago
cdk - CDK 2.9
Summary
- Improved abbreviation handling
- More arrow types
- Multi-step Reaction SMILES
- Reaction Set and Multi-step depiction
- More correct PubChemFingerprinter
- Universal (InChI) SMILES for large molecules
- Dependency updates and stability improvements, huge kudos to @uli-f for finding some longstanding issues
Improved abbreviation handling
991. The Abbreviation handling has been tweaked with more and cleaner options:
java
Abbreviations abbreviations = new Abbreviations();
// abbreviations.setContractToSingleLabel(true); // old (still supported)
abbreviations.with(Abbreviations.Option.ALLOW_SINGLETON); // new
// abbreviations.setContractOnHetero(true); // old (still supported)
abbreviations.with(Abbreviations.Option.AUTO_CONTRACT_HETERO); // new
The full options are described here: Abbreviations.Option.
More arrow types
Now includes NoGo/Equilibrium/RetroSynthetic - #927. See IReaction.Direction. Examples:
Multi-step Reaction SMILES
https://github.com/cdk/cdk/pull/986
An new entry point to the SMILES parser has been added to parse into a "multi-step" reaction where by the product of one step is the reactant the the next. The basic idea is to allow more than two '>'. Parts at even positions are reactants/products and odd positions are agents/catalysts/solvents.
Basic idea:
java
SmilesParser sp = new SmilesParser(SilentChemObjectBuilder.getInstance());
IReactionSet rset = sp.parseReactionSetSmiles("[Pb]>>[Ag]>>[Au] lead-to-silver-to-gold");
Real example (see next bullet for depiction):
ClC1=NC=2N(C(=C1)N(CC3=CC=CC=C3)CC4=CC=CC=C4)N=CC2C(OCC)=O>C1(=CC(=CC(=N1)C)N)N2C[C@H](CCC2)O.O1CCOCC1.CC1(C2=C(C(=CC=C2)P(C3=CC=CC=C3)C4=CC=CC=C4)OC5=C(C=CC=C15)P(C6=CC=CC=C6)C7=CC=CC=C7)C.C=1C=CC(=CC1)\C=C\C(=O)\C=C\C2=CC=CC=C2.C=1C=CC(=CC1)\C=C\C(=O)\C=C\C2=CC=CC=C2.C=1C=CC(=CC1)\C=C\C(=O)\C=C\C2=CC=CC=C2.[Pd].[Pd].[Cs]OC(=O)O[Cs]>C1(=CC(=CC(=N1)C)NC2=NC=3N(C(=C2)N(CC4=CC=CC=C4)CC5=CC=CC=C5)N=CC3C(OCC)=O)N6C[C@H](CCC6)O>CO.C1CCOC1.O.O[Li]>C1(=CC(=CC(=N1)C)NC2=NC=3N(C(=C2)N(CC4=CC=CC=C4)CC5=CC=CC=C5)N=CC3C(O)=O)N6C[C@H](CCC6)O>CN(C)C(=[N+](C)C)ON1C2=C(C=CC=N2)N=N1.F[P-](F)(F)(F)(F)F.[NH4+].[Cl-].CN(C)C=O.CCN(C(C)C)C(C)C>C1(=CC(=CC(=N1)C)NC2=NC=3N(C(=C2)N(CC4=CC=CC=C4)CC5=CC=CC=C5)N=CC3C(N)=O)N6C[C@H](CCC6)O>>C1(=CC(=CC(=N1)C)NC2=NC=3N(C(=C2)N)N=CC3C(N)=O)N4C[C@H](CCC4)O |f:4.5.6.7.8,16.17,18.19| US20190241576A1
Reaction Set and Multi-step depiction
https://github.com/cdk/cdk/pull/986
The DepictionGenerator has been extended to depict reaction sets. If the product of the previous reaction is the same as the reactant in the next (object identity) it is omitted for a terser depiction:
More correct PubChemFingerprinter
Explicit hydrogens are not longer required and there is an option to use a more correct ring set definition matching closer the original CACTVS substructure keys. This is now on by default:
IChemObject builder = SilentChemObjectBuilder.getInstance();
new PubchemFingerprinter(builder); // new - default is to use "ESSSR-like" ring set
new PubchemFingerprinter(builder, false); // old - for backwards compatible with FP generated with older CDK versions
Universal (InChI) SMILES for large molecules
979.
The InChI now supports > 999 atoms, we have the option to generate a SMILES using the InChI canonical labelling, it makes sense to use the larger molecules flag and support more.
New Contributors
- @Mailaender made their first contribution in https://github.com/cdk/cdk/pull/934
- @parit made their first contribution in https://github.com/cdk/cdk/pull/980
All Contributors
75 John Mayfield
17 Egon Willighagen
6 Uli Fechner
4 Mark J. Williamson
3 Mark Williamson
1 Parit Bansal
1 Matthias Mailänder
Full Changelog: https://github.com/cdk/cdk/compare/cdk-2.8...cdk-2.9
- Java
Published by johnmay over 2 years ago
cdk - CDK 2.8
Key Changes
- JDK Versions:
- JDK 8 (minimum)
- JDK 11 (minimum if not using cdk-iordf)
- JDK 17 (recommended)
- The project is now built with Java 11+ but compiled to target Java 8. If you have any issues please let us know.
- The master branch has been renamed to main.
- logj4-core is no longer a dependency of cdk-log4j, you should include these separately if you intend to use Log4j
- A new cdk-slf4j module allows connecting logging to SLF4J
- MayGen structure generator, provides the ability to generate millions and millions of structures that have a given formulae.
java Maygen maygen = new Maygen(SilentChemObjectBuilder.getInstance()); maygen.setFormula("C3Cl2H4"); maygen.setConsumer(new SmiOutputConsumer(new OutputStreamWriter(System.out))); maygen.run();Maygen is pure Java, if you need more speed consider Surge by the same author. - New Smallest Ring utilities for single atom/bond
java if (Cycles.smallRingSize(atom, 7) != 0) { // atom is in a ring 7 or smaller } if (Cycles.smallRingSize(bond, 7) != 0) { // bond is in a ring 7 or smaller } - RAW/Count Path Fingerprints
java IFingerprinter fpr = new Fingerprinter(); Map<String, Integer> feats = fpr.getRawFingerprint(mol); Where possible "Re-inflate" convex rings on cyclcophanes: Before:
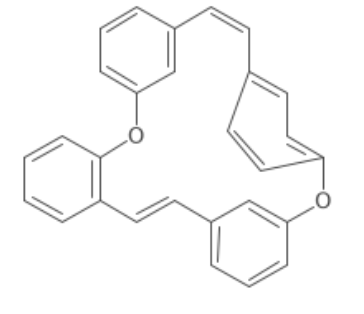 now:
now: 
New substructure/copy utility that allows a whole or part of a structure to be copied. Atoms are bonds are selected by providing a predicate: ```java IAtomContainer dst = builder.newAtomContainer(); AtomContainerManipulator.copy(dst, src, a -> a.isInRing(), b -> b.isInRing()); // select the cyclic part of a molecule
// select atoms in a set, the bonds will also be selected
Set
- New *exclusive* atoms filter that provides non-overlapping substructure matches, note the input order can determine which matches are selected.
java
for (int[] mapping : Pattern.findSubstructure(query).matchAll(mol).exclusiveAtoms()) {
// ...
}
```
- Stereo perception corner-cases. Reject:
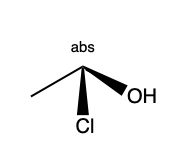 ,
,
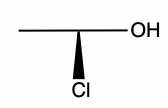 , ok:
, ok:
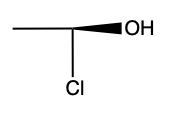
Summary
- Merged all PRs and resolved all open issues related to bugs
- InChINumbersTools: Use JNA InChI options by @johnmay in https://github.com/cdk/cdk/pull/799
- Avoid integer overflow in MF by @johnmay in https://github.com/cdk/cdk/pull/808, https://github.com/cdk/cdk/pull/810
- Ensure correct stereo consistency (Fix #812) by @johnmay in https://github.com/cdk/cdk/pull/813
- SMILES: Fix an issue with stereochemistry being lost on generic atoms - @johnmay in https://github.com/cdk/cdk/pull/814, https://github.com/cdk/cdk/pull/866
- Maygen structure generator by @MehmetAzizYirik in https://github.com/cdk/cdk/pull/811
- Weighted path descriptor performance improvements by @johnmay in https://github.com/cdk/cdk/pull/817
- Depiction: Fix missing bond annotations by @johnmay in https://github.com/cdk/cdk/pull/819
- Utility functions for determining the smallest ring size of an atom/b… by @johnmay in https://github.com/cdk/cdk/pull/820
- Better consistentcy in Stereochemistry and Sgroups when removing atoms by @johnmay in https://github.com/cdk/cdk/pull/821
- Unify MOLfile V2000/V3000 options by @johnmay in https://github.com/cdk/cdk/pull/824, https://github.com/cdk/cdk/pull/852
- Improved stereochemistry perception by @johnmay in https://github.com/cdk/cdk/pull/826, https://github.com/cdk/cdk/pull/839
- Replace Atom symbol (String) comparison with atomic number (integer) by @johnmay in https://github.com/cdk/cdk/pull/827
- Improved/fix bugs with XLogP, PiContact, and BCUT, HuLuIndex descriptors by @johnmay in https://github.com/cdk/cdk/pull/833, https://github.com/cdk/cdk/pull/656, https://github.com/cdk/cdk/pull/822, https://github.com/cdk/cdk/pull/832
- Additional Raw and count path fingerprints by @johnmay in https://github.com/cdk/cdk/pull/834
- "Re-inflate" convex rings in macrocycles. The macrocycle layout can en… by @johnmay in https://github.com/cdk/cdk/pull/836
- Fix a corner case in repeat crossing bonds when we have variable atta… by @johnmay in https://github.com/cdk/cdk/pull/835
- Restore space as delimiter for string-based definition of InChI options by @marco-foscato in https://github.com/cdk/cdk/pull/846
- Update to Apache Jena 4.2 (requires JDK 11) by @egonw in https://github.com/cdk/cdk/pull/748
- Fix localisation of alpha channel floats in SVG by @egonw in https://github.com/cdk/cdk/pull/868
- Check string bounds on PDB COMPND line. Fixes #870 by @johnmay in https://github.com/cdk/cdk/pull/871
- Methods to manipulate atom types in ReactionManipulator by @uli-f in https://github.com/cdk/cdk/pull/883, https://github.com/cdk/cdk/pull/879
- Added ChemObjectBuilder.newReaction() by @uli-f in https://github.com/cdk/cdk/pull/888
- Utilities for selecting a substructure of a molecule. by @johnmay in https://github.com/cdk/cdk/pull/889
- Improved CDK Log4J/SLF4J interactions by @johnmay in https://github.com/cdk/cdk/pull/878, https://github.com/cdk/cdk/pull/876
- Additional SMARTS/matching utilities by @johnmay in https://github.com/cdk/cdk/pull/896, https://github.com/cdk/cdk/pull/900
- Use Junit5 by @johnmay in https://github.com/cdk/cdk/pull/901
- Fix issue with hose code nesting by @johnmay in https://github.com/cdk/cdk/pull/828
Authors
278 John Mayfield
13 Egon Willighagen
11 Uli Fechner
5 Mark Williamson
3 Valentyn Kolesnikov
2 MehmetAzizYirik
2 Marco Foscato
1 dependabot[bot]
1 Tim Dudgeon
1 Otto Brinkhaus
1 Christoph Steinbeck
1 Alex
New Contributors
- @marco-foscato made their first contribution in https://github.com/cdk/cdk/pull/846
- @tdudgeon made their first contribution in https://github.com/cdk/cdk/pull/847
- @OBrink made their first contribution in https://github.com/cdk/cdk/pull/851
- @sashashura made their first contribution in https://github.com/cdk/cdk/pull/885
Full Changelog: https://github.com/cdk/cdk/compare/cdk-2.7.1...cdk-2.8
- Java
Published by johnmay over 3 years ago
cdk - CDK 2.7.1
This page documents the changes for CDK v2.7 and v2.7.1. The patch version was made after some minor issues with how the new InChI code was organised were discovered by downstream projects.
Features
Switch from JNI to JNA InChI.
There are two main technologies for calling native code JNI (Java Native Interface) and JNA (Java Native Access). JNI requires writing a custom native wrapper which is then bound to Java code, JNA allows you to call the native methods of an existing SO/DYLIB directly. Essentially what this means is to expose the native InChI library in Java one needs to first write (and maintain) a native wrapper, with JNA we can just drop the InChI SO directly in. JNI InChI exposed InChI v1.03 and worked well for many years - unfortunately this project was no longer maintained and as newer more stable versions of InChI were released (now v1.06) an alternative was needed. A few years ago Daniel Lowe started JNA InChI and recently made it feature complete and released v1.0.
ChemAxon have also independently used the JNA path to integrated newer InChI libraries into their tools: (slides). It is not clear if this was made available, it is not listed on GitHub/ChemAxon.
Build on Java 17
The Maven plugins were updated to allow building on Java 17
Verify declared dependencies
The maven modules were checked for unused declared dependencies and used undeclared dependencies (mvn dependency:analyze).
Organise and restructure test-jar and testdata
CDK was originally built with the ant build tool, under this scheme there was a jar for the main/ code and one the test/ code. Test modules could share an inherit dependencies. To replicate this in maven we install and deploy "test-jar" artefacts. The project test code was restructured to put all common test code in the "cdk-test" module.
All test data was stored in a cdk-testdata module, this data has now been relocated to the test/resources of each module where it is used. This meant some data was duplicated but means the ~18MB test-jar no longer needs to be uplodaded to maven central.
Remove Guava dependency
We have removed the use of Guava, the functionality could mostly be directly replaced with newer JDK idioms (Function/Predicate/Stream) which were not available in the past.
Use XorShift PRNG in ShortestPathFingerprinter (different fingerprint)
Commons Math3 was used in a single place to hash paths (Mersenne Twister) in the ShortestPathFingerprinter. Since this fingerprint method is not widely used and the hashes do not need to be cryptographically secure a simple https://en.wikipedia.org/wiki/Xorshift random generate is now used instead. This allows us to remove the dependency on Commons Math3. This does mean the fingerprint bits have changed, note the CDK version description is accessible via the Fingerprinter.getVersionDescription() method.
Authors
137 John Mayfield
6 Egon Willighagen
1 dependabot[bot]
Full Change Log
- Bump version ready for development John Mayfield on 2021-12-14
- Bumped the log4j version Egon Willighagen on 2021-12-14
- Make log4j a test-only dependency of the InChI module John Mayfield on 2021-12-16
- Make sure TOTAL_DEGREE works correctly John Mayfield on 2021-12-16
- Additional test for converting "Molecules" to queries using the useful SMARTS therom: D=X+h. John Mayfield on 2021-12-16
- Updated CMLXOM version Egon Willighagen on 2021-12-16
- Update Mockito to 4.1.0 - fixing test failures (except InChI) on M1 AARCH64. John Mayfield on 2021-12-16
- Add in a Java 17 build John Mayfield on 2021-12-16
- Fix indents John Mayfield on 2021-12-16
- Update Jacoco plugin John Mayfield on 2021-12-16
- Formatting only - to make changes easier to follow John Mayfield on 2021-12-18
- First pass at moving from JNA to JNI inchi - some tests need adjusting. John Mayfield on 2021-12-18
- Fix Test - ignore longer extended tetrahedral - could be a warning. John Mayfield on 2021-12-18
- 5000L (long) default timeout in ms John Mayfield on 2021-12-18
- This is not a status message rather than log - makes sense John Mayfield on 2021-12-18
- These tests add the same bond twice a1E=a2E which is now a warning - used to be ignored. The tests were wrong John Mayfield on 2021-12-18
- This is now a warning, there is no EOF status. However it should perhaps set a sensible message John Mayfield on 2021-12-18
- More double bonds added twice. John Mayfield on 2021-12-18
- Message is empty string rather than null. John Mayfield on 2021-12-18
- This is the most questionable change but believe to be a bug in InChI 1.03. Using JNI INCHI setting the chiral flag = on or off we get "rA:9n..." without it we get ""rA:9...". In JNA INCHI we always get "rA:9n..." - this molecule is not chiral so it seems odd that the setting would change anything. Since this is only a change in AuxInfo this is acceptable. John Mayfield on 2021-12-18
- Looks like we can different timeout messages based on the system? John Mayfield on 2021-12-18
- Bump log4j-core from 2.16.0 to 2.17.0 dependabot[bot] on 2021-12-18
- Some minor version/scope cleanups. Hamcrest should be a test dependency. Make sure we pull in Log4j2 2.17.0. In QSARCML log4j-core should only be in the tests John Mayfield on 2021-12-22
- Move over to Log4J2 configuration - allows us to remove some log4j-1.2 deps John Mayfield on 2021-12-22
- We don't need the Log4J 1.2 API in these locations - unfortunately it still comes in via CMLXOM and JENA but better for now. John Mayfield on 2021-12-22
- Cleanup of the cdk/base modules - using dependency analzye to ensure all used undelcared dependencies are included and unused declared are removed. John Mayfield on 2021-12-22
- Cleanup of dependencies in CDK storage/io modules John Mayfield on 2021-12-22
- Significant dependency cleanup in the descriptor/qsar modules. John Mayfield on 2021-12-22
- Cleanup dependencies in depict/render module John Mayfield on 2021-12-22
- Cleanup dependencies in CDK tool modules. John Mayfield on 2021-12-22
- Cleanup dependencies in the misc/ module John Mayfield on 2021-12-22
- Broken by changes to another module - it implicitly depended on the CDK atomtyping. John Mayfield on 2021-12-22
- Make sure everything is used is declared in the cdk-legacy module John Mayfield on 2021-12-22
- More cleanup of base/ modules now I've got better at using dependency:analyze John Mayfield on 2021-12-22
- Minor issues of non-test dependencies now a clean build is tested John Mayfield on 2021-12-22
- More left overs - all good now. John Mayfield on 2021-12-22
- This should probably be install instead of test John Mayfield on 2021-12-22
- Looks like some things were folded into JDK 17 John Mayfield on 2021-12-22
- JENA-CORE pulls in a very specific version XML-APIS. There may well be a conflict but a fix should be to leave it as a transient dependency in cdk-io John Mayfield on 2021-12-22
- Avoid test-jar dependency for qsarcml - we only need the roundtrip function. John Mayfield on 2021-12-24
- This code is deprecated we don't need the full Desctiptor basic checks. Note it may make sense to move the descriptor interfaces to CDK interfacts then the descriptor tests can go to into cdk-test. John Mayfield on 2021-12-24
- Duplicate some basic generator tests. John Mayfield on 2021-12-24
- We can from cdk-fingerprint -> cdk-test-standard:test-jar:test by moving the required classes to where they are needed (they are not needed anywhere else). John Mayfield on 2021-12-24
- Remove cdk-diff dependency on cdk-test, it only used very basic assertion utilites which we can just write out or duplicate. John Mayfield on 2021-12-24
- Now cdk-diff is essenitaly independant we can completely from the cdk-test-inferfaces module and move all of the abstract tests into the main cdk-test. John Mayfield on 2021-12-24
- Eliminate cdk-test-core dependencies by moving some helper classes to cdk-test John Mayfield on 2021-12-24
- Move the TestMoleculeFactory to cdk-data/main. I'm not super happy about this but seems like the simplest solution. The other being to make the tests pass in a builder, or have the test molecule factory dynamically find the IChemObjectBuilder - that would be good except for now Java really doesn't like reflective access between modules. John Mayfield on 2021-12-24
- First of let's ignore some trivial tests that check a ChemFile etc is accepted... will we work out how to add these back in once relocated but the tests aren't THAT useful really. John Mayfield on 2021-12-24
- Demonstrates the tests are perhaps a little odd John Mayfield on 2021-12-24
- Duplicate the expectReader test from the FormatFactory. John Mayfield on 2021-12-24
- Extract out the interfaces for Rgroup queries so we can beter seperate tests. The Rgroup queries need a rethink in my opinion but this will do for now. John Mayfield on 2021-12-24
- We will be mocking this IChemModel and so no builder will be avaliable - rewrite to avoid creating IAtomContainerSets (and a temp ChemModel). John Mayfield on 2021-12-24
- Mock the ChemObjectIO test cases John Mayfield on 2021-12-24
- Remove test-jar dependencies on cdk-qsar. We can relocate the required interfaces to cdk-interfaces and add an optional setDescriptor method that takes a builder. We can then move it to cdk-test. John Mayfield on 2021-12-24
- One more in cdk-legacy John Mayfield on 2021-12-24
- Duplicate MolecularDescriptorTest for qsarprotein - I would actually vote to just move the two qsarprotein tests into qsarmolecular John Mayfield on 2021-12-24
- Need this dependency since scope=test doesn't pull in the extras John Mayfield on 2021-12-24
- Eliminate testdata dependency on cdk-charges. John Mayfield on 2021-12-29
- Eliminate testdata from cdk-sdg moving all resources to the module (and package) where they are used. John Mayfield on 2021-12-29
- Eliminate testdata from cdk-builder3d moving all resources to the module (and package) where they are used. We need to duplicate one resource also used in cdk-io test. John Mayfield on 2021-12-29
- Eliminate testdata from cdk-formula moving all resources to the module (and package) where they are used. John Mayfield on 2021-12-29
- Eliminate testdata from cdk-pcore moving all resources to the module (and package) where they are used. John Mayfield on 2021-12-29
- Relocate required file from testdata to where it is needed. John Mayfield on 2021-12-29
- Generics fixup John Mayfield on 2021-12-29
- forcefield doesn't need testdata John Mayfield on 2021-12-29
- cdk-qsarcml, cdk-qsarbond and cdk-qsarprotein do not need testdata John Mayfield on 2021-12-29
- Eliminate cdk-cip dependency on testdata, one dupe needed John Mayfield on 2021-12-29
- Eliminate cdk-qsaratomic dependance on testdata John Mayfield on 2021-12-29
- Seperate cdk-qsarmolecular from testdata, 4 files need duplicating to other modules as well. John Mayfield on 2021-12-29
- Seperate out cdk-fingerprint from testdata, 2 test files needed duplicating. John Mayfield on 2021-12-29
- Some collatoral damage - there is a "defeat device" that tries to slurp up all MDL files and run them though the atom typer. Probably will just replace with SMILES or at best an SDFfile. For now let's keep it working as we expect. John Mayfield on 2021-12-29
- Split out cdk-legacy from testdata - most of the dupes here are for that one atomtype matcher test John Mayfield on 2021-12-29
- Eliminate testdata from test-valencycheck John Mayfield on 2021-12-29
- Seperate out testdata from iordf. John Mayfield on 2021-12-29
- Eliminate testdata dependency from cdk-inchi. John Mayfield on 2021-12-29
- Eliminate the testdata dependency from PDB John Mayfield on 2021-12-29
- pdbcml is now independent of testdata - due to cdk-pdb changes. John Mayfield on 2021-12-29
- Eliminate testdata dependency from cdk-libiocml John Mayfield on 2021-12-29
- Eliminate testdata dependency from cdk-smiles. As before most of the duplicatations are due to the cdk-core AtomType "MDLfiles" test John Mayfield on 2021-12-29
- Eliminate testdata dependency from cdk-reaction John Mayfield on 2021-12-29
- Eliminate testdata dependency from cdk-ctab. John Mayfield on 2021-12-29
- Eliminate testdata dependency from cdk-ioformats, we need some duplication in core/extra an io. John Mayfield on 2021-12-29
- Eliminate testdata dependency from cdk-io. John Mayfield on 2021-12-29
- Eliminate testdata dependency from test-extra. John Mayfield on 2021-12-29
- Eliminate testdata dependency from cdk-test-atomtype John Mayfield on 2021-12-29
- Eliminate testdata dependency from cdk-test-core. John Mayfield on 2021-12-29
- Eliminate testdata dependency from cdk-test-standard. John Mayfield on 2021-12-29
- The explicit testdata module can now be deleted, all files have been moved where they are needed - remaining files are not used. Note some were copied i.e. for the "mdlFiles" CDKAtomTypeMatcher test. John Mayfield on 2021-12-29
- Move cdk-test test/ code to main/ - we now expect the modules using it should include via scope=test. John Mayfield on 2021-12-29
- We no longer need to build test-jars as none are be used. This saves the additional uploads to central. John Mayfield on 2021-12-29
- Bumped log4j to 2.17.1 Egon Willighagen on 2021-12-30
- Bumped CMLXOM to the latest version Egon Willighagen on 2021-12-31
- We can replace checkNotNull with requireNonNull (since JDK 1.7) John Mayfield on 2022-01-01
- There is no replacement for checkArgument in recent JDK's - however the logic is simple enough that we can just expand out the conditions. John Mayfield on 2022-01-01
- IDE warns about Jacoco version missing from sub-modules (e.g. test-standard) - let's define it once in the parent. John Mayfield on 2022-01-01
- Cleanup some other pom issues reported by the IDE, prerequisites is for plugins, cdk-qsar was duplicated John Mayfield on 2022-01-01
- Replace Guava Predicate/Function with the JDK 1.8 versions. This technically breaks the API but is very easy to update thanks to the :: operator. We will be revisiting the Mappings.java to use the Streams API John Mayfield on 2022-01-01
- Use JDK 1.7 Objects methods John Mayfield on 2022-01-01
- JDK 1.8 has Charsets John Mayfield on 2022-01-01
- Joiner -> String.join, since JDK 1.8 John Mayfield on 2022-01-01
- This was a useful one, the "official" calculation is (int)1+(n/0.75) for the default load factor. However in all these cases we expected n to be small - say 100 or so. In which case 135 vs 200 (2*n) is fine. John Mayfield on 2022-01-01
- Fixup - Objects John Mayfield on 2022-01-01
- Replace the XmlEscape from Guava, this was a unstable API anyways. We really just need >/< to be replaced but have handled the control characters anyways. John Mayfield on 2022-01-01
- Expected HashSet size John Mayfield on 2022-01-01
- Avoid Longs.toArray and just create the array John Mayfield on 2022-01-01
- Replace Guava CharStreams with normal JDK 1.8 streams John Mayfield on 2022-01-01
- Remove Guava caches, one is no longer needed - we have matchRoot now. The second case is technically incorrect as the contents of the IAtomContainer may change so it's not safe to cache here. There is a bug with AtomContainer2.add which will be fixed in another commit - for now we can use the copy constructor. John Mayfield on 2022-01-01
- Replace Lists usage - diamond brackets (1.7+) make thinks more concise than the old help function. John Mayfield on 2022-01-01
- Replace usage of Guava Ints utility. Integer.compare helps as well as IntStream John Mayfield on 2022-01-01
- Replace usage of Guava's BiMap John Mayfield on 2022-01-01
- ImmutableSet removal John Mayfield on 2022-01-01
- Replace usages of MultiMap - the computeIfAbsent and getOrDefault APIs of Map now make this construct on vanialla JDK easy. We need a slight tweak to our Cycle comparator since TreeMulitmap keeps things sorted by keys and then values. Since the key is just the length we can keep things equivalent by first comparing the length. John Mayfield on 2022-01-01
- We can now use Java 8 streams to filter an iterator - more verbose. John Mayfield on 2022-01-01
- Remove usage of FluentIterable using the Stream API. Some usages are more verbose but OK and perhaps indicate a List should have been consumed/returned in the first place. John Mayfield on 2022-01-01
- Replace Iterables and Iterables usage - alot of this can be handled with Stream's now. Again slightly more verbose, there is one extra case which needs special comment due to mocking. John Mayfield on 2022-01-01
- Here we do the old-school iteration, we canot call stream().count() because it use the forEachRemaining method which will was not mocked. John Mayfield on 2022-01-01
- Improved Mocking to allow use to use the stream().count() method. John Mayfield on 2022-01-01
- Replace ImmutableMap usage, JDK 9+ has Map.of() which is a direct replacement. However usage is minimal so we can just do it the verbose way. John Mayfield on 2022-01-01
- Remove Guava dependency John Mayfield on 2022-01-01
- Add a test of the HETATM atom types. John Mayfield on 2022-01-03
- Compress the PDB hetatm type_map by not storing the common C.sp2 and H types. John Mayfield on 2022-01-03
- Sort the type_map.txt lexicographically for a slightly smaller JAR footprint - add a comment explaining the common type handling. John Mayfield on 2022-01-03
- We need to also handle residues that are 100% C.sp2 and H John Mayfield on 2022-01-03
- Bumped to CMLXOM 4.0 Egon Willighagen on 2022-01-03
- We don't actually use FreeHEP to SVG output anymore - we have our own SVG draw visitor since it's such a simple format. John Mayfield on 2022-01-03
- Eliminate commons-math3 dependency by reimplementing the MersenneTwister logic. This was only used for a over-complicated PRNG used in the shortest path fingerprinter. A simpler linear-feedback shift PRNG would work just as well here but we want to keep backwards compatbility. Note this fingerprint it's really very good since the fingerprints are non-transativie and can't be used for substructure screening. John Mayfield on 2022-01-04
- Use a simpler and faster random number generate to hash fingerprint bits. Note we actually set more bits in a test fingerprint so in that particualar case the features were better distributed. John Mayfield on 2022-01-04
- Better comment John Mayfield on 2022-01-04
- Fix HTML5 JavaDoc errors (JDK 17). John Mayfield on 2022-01-06
- cdk-test code is now in the main/ so goes through the JavaDoc (will fix later) - but we should make sure it doesn't have errors in it. First off there many case of a retrun on a void - looks like a copy paste error. John Mayfield on 2022-01-06
- Some others errors in cdk-test John Mayfield on 2022-01-06
- Relocate all classes in cdk-test to the package org.openscience.cdk.test.*. John Mayfield on 2022-01-07
- Exclude anything in org.openscience.cdk.test from the JavaDoc John Mayfield on 2022-01-07
- Latest version of JavaDoc plugin John Mayfield on 2022-01-07
- CDK 2.7 John Mayfield on 2022-01-08
- New dev version 2.8-SNAPSHOT John Mayfield on 2022-01-09
- Update README John Mayfield on 2022-01-09
- Use HTTPS for EBI plugin repo John Mayfield on 2022-01-09
- Move the old JNI enums to a seperate (optional) module - this means the InChI works fine. If you need the enums you can optionally include the cdk-jniinchi-support module. John Mayfield on 2022-01-10
- Include in the cdk-bundle by default John Mayfield on 2022-01-10
- Deprecate the JNI Inchi return/inputs and provide preferred JNA inchi alternatives. John Mayfield on 2022-01-10
- Match the version in master Egon Willighagen on 2022-01-10
- Make sure we have correct storage order in a bond when reading from InChI. John Mayfield on 2022-01-11
- Version 2.7.1 John Mayfield on 2022-01-11
- Java
Published by johnmay almost 4 years ago
cdk - CDK 2.2
Please see 2.2 Release Notes for full details.
- Java
Published by johnmay about 7 years ago
cdk - CDK 2.1.1 (patch release)
This patch release removes the SNAPSHOT dependency. Release Notes
- Java
Published by johnmay about 8 years ago
cdk - CDK Release 1.5.14
https://github.com/cdk/cdk/wiki/1.5.14-Release-Notes
- Java
Published by johnmay about 9 years ago
cdk - CDK Release 1.5.11
https://github.com/cdk/cdk/wiki/1.5.11-Release-Notes
- Java
Published by johnmay over 10 years ago
cdk - CDK Release 1.5.10
https://github.com/cdk/cdk/wiki/1.5.10-Release-Notes
- Java
Published by johnmay about 11 years ago