rapidgzip
Gzip Decompression and Random Access for Modern Multi-Core Machines
Science Score: 67.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
✓CITATION.cff file
Found CITATION.cff file -
✓codemeta.json file
Found codemeta.json file -
✓.zenodo.json file
Found .zenodo.json file -
✓DOI references
Found 5 DOI reference(s) in README -
✓Academic publication links
Links to: arxiv.org -
○Committers with academic emails
-
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (12.9%) to scientific vocabulary
Keywords
Repository
Gzip Decompression and Random Access for Modern Multi-Core Machines
Basic Info
Statistics
- Stars: 429
- Watchers: 7
- Forks: 12
- Open Issues: 8
- Releases: 1
Topics
Metadata Files
README.md
[](https://github.com/mxmlnkn/rapidgzip/blob/main/CHANGELOG.md) [](http://opensource.org/licenses/MIT) [](https://github.com/mxmlnkn/rapidgzip/actions/workflows/test-python.yml)  [](https://discord.gg/Wra6t6akh2) [](https://t.me/joinchat/FUdXxkXIv6c4Ib8bgaSxNg) 
This repository contains the command line tool rapidgzip, which can be used for parallel decompression of almost any gzip file. Other tools, such as bgzip, can only parallelize decompression of gzip files produced by themselves. rapidgzip works with all files, especially those produced by the usually installed GNU gzip. How this works can be read in the pugz paper or in the rapidgzip paper, which builds upon the former.
The Python module provides a RapidgzipFile class, which can be used to seek inside gzip files without having to decompress them first.
Alternatively, you can use this simply as a parallelized gzip decoder as a replacement for Python's builtin gzip module in order to fully utilize all your cores.
The random seeking support is the same as provided by indexed_gzip, but further speedups are realized at the cost of higher memory usage thanks to a least-recently-used cache in combination with a parallelized prefetcher.
This repository is a light-weight fork of the indexed_bzip2 repository, in which the main development takes place. This repository was created for visibility reasons and in order to keep indexed_bzip2 and rapidgzip releases and packaging separate. It will be updated at least for each release. Issues regarding rapidgzip should be opened here.
Table of Contents
Installation
You can simply install it from PyPI:
python3 -m pip install --upgrade pip # Recommended for newer manylinux wheels
python3 -m pip install rapidgzip
rapidgzip --help
Advanced Installations
To build from source, these packages are necessary (Debian and derivatives): ```bash sudo apt install git gcc g++ python3 python3-dev python3-pip python3-build python3-venv nasm ``` Then, the latest unreleased development version can be built with: ```bash python3 -m pip install --force-reinstall 'git+https://github.com/mxmlnkn/rapidgzip.git@main#egginfo=rapidgzip&subdirectory=python/rapidgzip' ``` And to build locally, you can use `build` and install the wheel: ```bash git clone --recursive https://github.com/mxmlnkn/rapidgzip.git cd rapidgzip/python/rapidgzip/ rm -rf dist python3 -m build python3 -m pip install --force-reinstall --user --break-system-packages dist/rapidgzip-*.whl ```Performance
Following are benchmarks showing the decompression bandwidth over the number of used cores.
There are two rapidgzip variants shown: (index) and (no index).
Rapidgzip is generally faster when given an index with --import-index because it can delegate the decompression to ISA-l or zlib while it has to use its own custom-written gzip decompression engine when no index exists yet.
Furthermore, decompression can be parallelized more evenly and more effectively when an index exists because the serializing window propagation step is not necessary.
The violin plots show 20 repeated measurements as a single "blob". Thin blobs signal very reproducible timings, while thick blobs signal a large variance.
Scaling Benchmarks on 2xAMD EPYC CPU 7702 (2x64 cores)
Decompression of Silesia Corpus
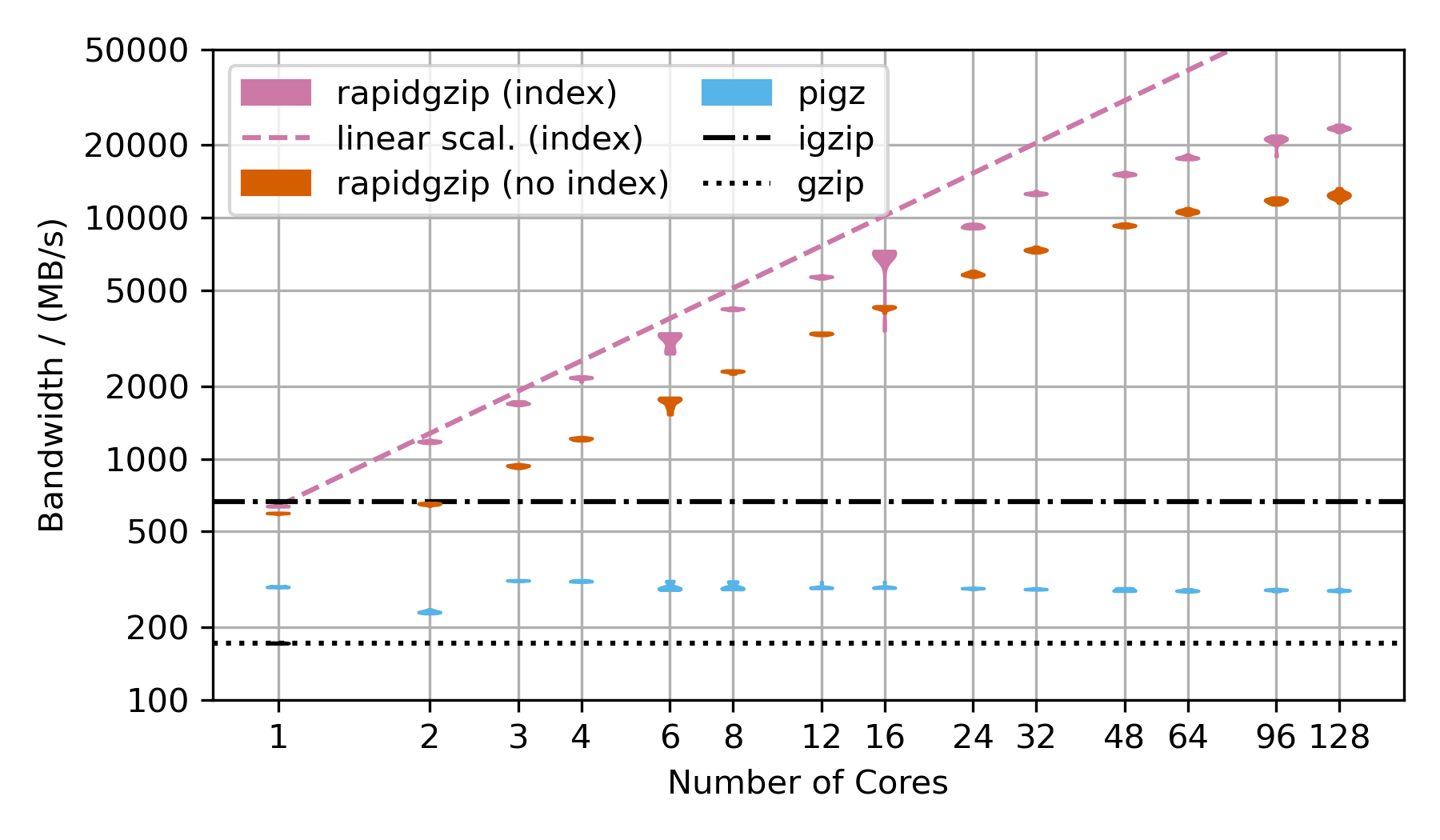
This benchmark uses the Silesia corpus compressed as a .tar.gz file to show the decompression performance. However, the compressed dataset is only ~69 MB, which is not sufficiently large to show parallelization over 128 cores. That's why the TAR file is repeated as often as there are number of cores in the benchmark times 2 and then compressed into a single large gzip file, which is ~18 GB compressed and 54 GB uncompressed for 128 cores.
Rapidgzip achieves up to 24 GB/s with an index and 12 GB/s without.
Pugz is not shown as a comparison because it is not able to decompress the Silesia dataset because it contains binary data, which it cannot handle.
More Benchmarks
### Decompression of Gzip-Compressed Base64 Data 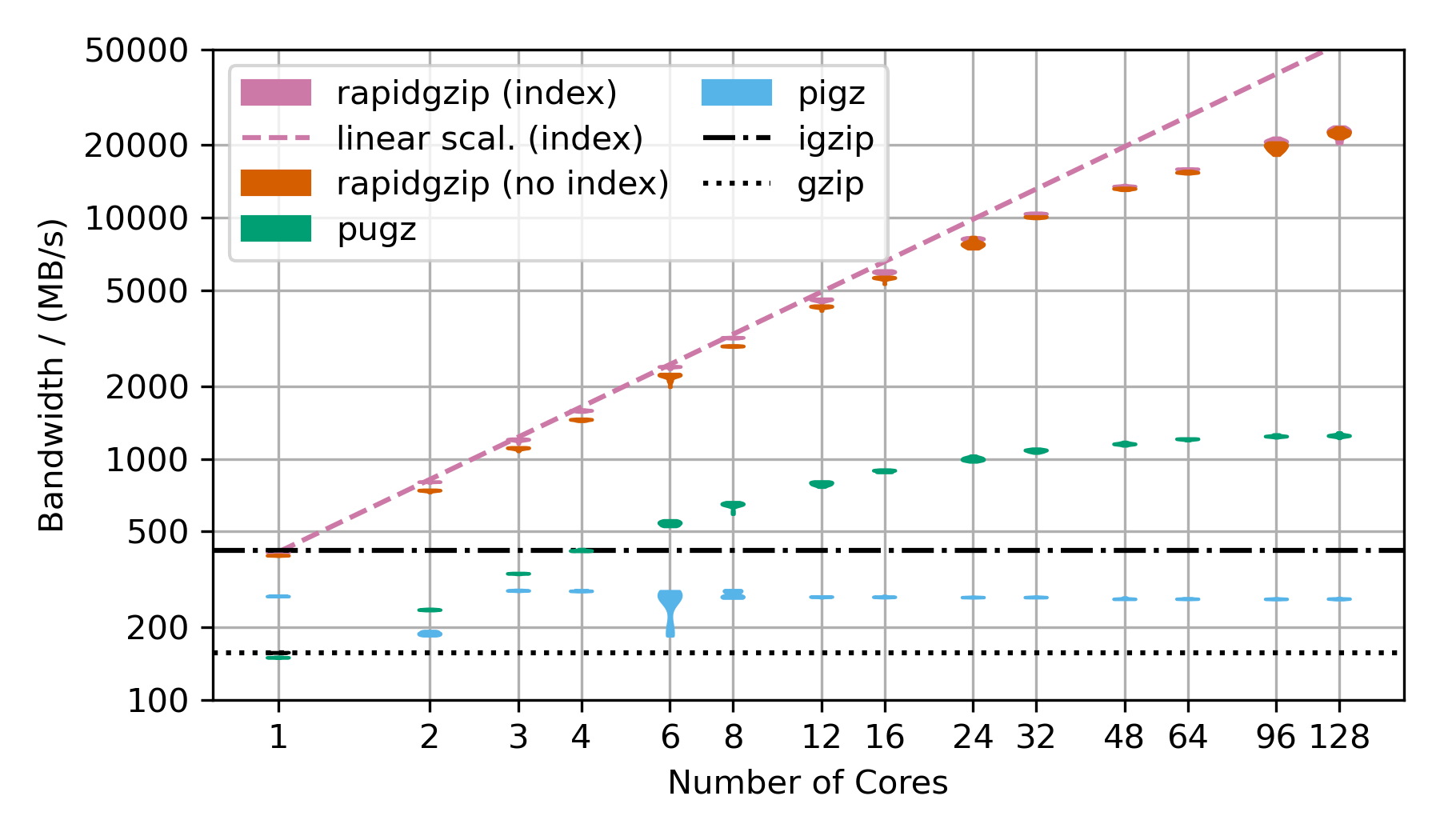 This benchmark uses random data, that has been base64 encoded and then gzip-compressed. This is the next best case for rapidgzip after the trivial case of purely random data, which cannot be compressed and therefore can be decompressed with a simple memory copy. This next best case results in mostly Huffman-coding compressed data with only very few LZ77 back-references. Without LZ77 back-references, parallel decompression can be done more independently and therefore faster than in the case of many LZ77 back-references. ### Decompression of Gzip-Compressed FASTQ Data 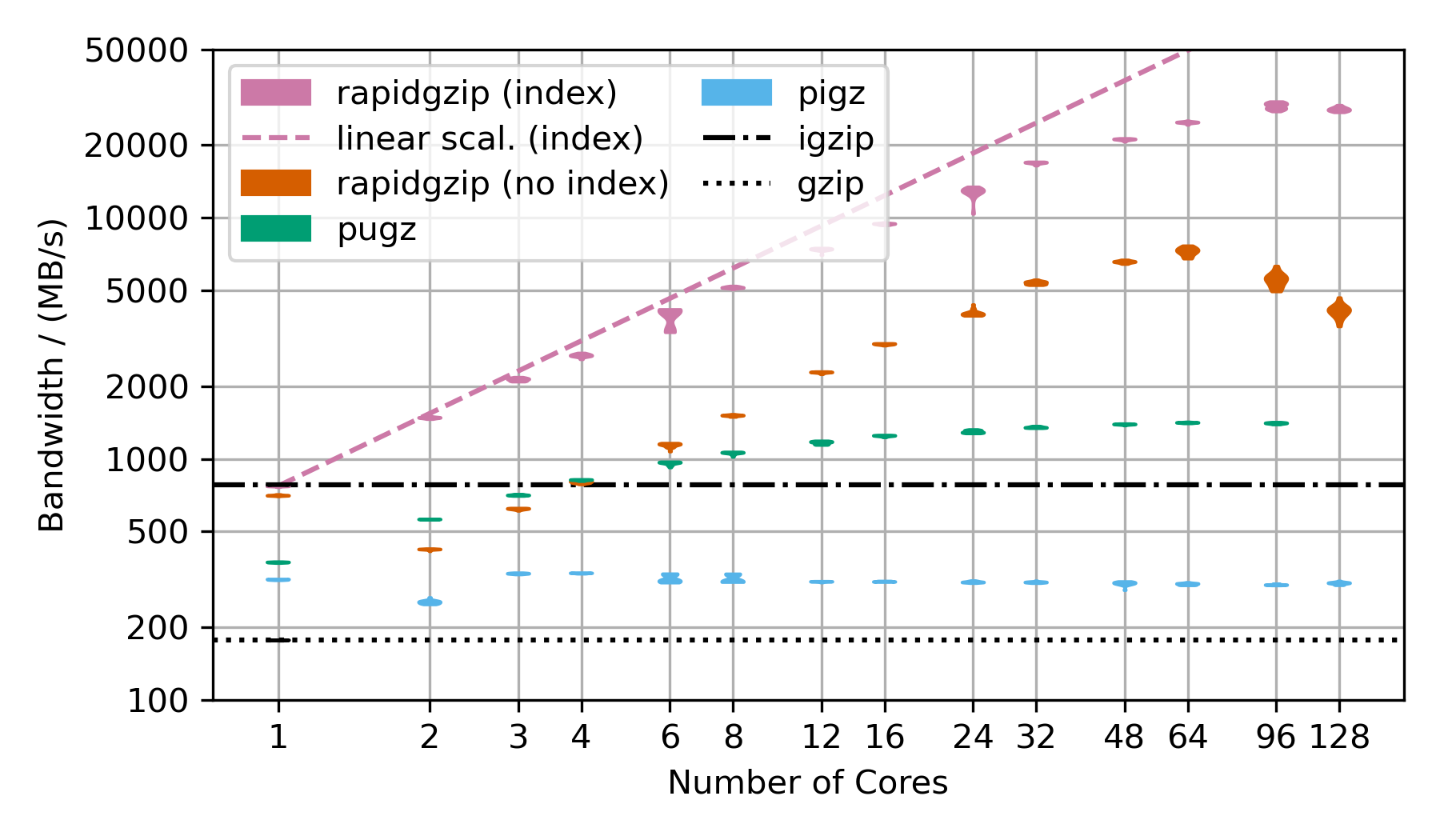 This benchmarks uses gzip-compressed [FASTQ data](http://ftp.sra.ebi.ac.uk/vol1/fastq/SRR224/085/SRR22403185/SRR22403185_2.fastq.gz). That's why the TAR file is repeated as often as there are number of cores in the benchmark to hold the decompression times roughly constant in order to make the benchmark over this large a range feasible. This is almost the worst case for rapidgzip because it contains many LZ77 back-references over very long ranges. This means that a fallback to ISA-L is not possible and it means that the costly two-staged decoding has to be done for almost all the data. This is also the reason why it fails to scale above 64 cores, i.e, to the second CPU socket. The first and second decompression stages are completely independently submitted to a thread pool, which on this NUMA architecture means, that data needs to be costly transferred from one processor socket to the other if the second step for a chunk is not done on the same processor as the first. This should be fixable by making the ThreadPool NUMA-aware. These three scaling plots were created with rapidgzip 0.9.0, while the ones in the [paper](https://github.com/mxmlnkn/indexed_bzip2/blob/master/results/paper/Knespel%2C%20Brunst%20-%202023%20-%20Rapidgzip%20-%20Parallel%20Decompression%20and%20Seeking%20in%20Gzip%20Files%20Using%20Cache%20Prefetching.pdf) were created with 0.5.0. ## Scaling Benchmarks on Ryzen 3900X These benchmarks on my local workstation with a Ryzen 3900X that only has 12 cores (24 virtual cores), but the base frequency is much higher than the 2xAMD EPYC 7702 CPU. ### Decompression With Existing Index | | 4GiB-base64 | 4GiB-base64 | | 20x-silesia | 20x-silesia |------------------------|------------------------------|-----------------|-|-------------------------------|--------- | Uncompressed Size | 4 GiB | | | 3.95 GiB | | Compressed Size | 3.04 GiB | | | 1.27 GiB | | **Module** | **Bandwidth/ (MB/s)** | **Speedup** | | **Bandwidth
/ (MB/s)** | **Speedup** | gzip | 250 | 1 | | 293 | 1 | rapidgzip (0 threads) | 5179 | 20.6 | | 5640 | 18.8 | rapidgzip (1 threads) | 488 | 1.9 | | 684 | 2.3 | rapidgzip (2 threads) | 902 | 3.6 | | 1200 | 4.0 | rapidgzip (6 threads) | 2617 | 10.4 | | 3250 | 10.9 | rapidgzip (12 threads) | 4463 | 17.7 | | 5600 | 18.7 | rapidgzip (24 threads) | 5240 | 20.8 | | 5750 | 19.2 | rapidgzip (32 threads) | 4929 | 19.6 | | 5300 | 17.7 ### Decompression From Scratch | | 4GiB-base64 | 4GiB-base64 | | 20x-silesia | 20x-silesia |------------------------|------------------------------|-----------------|-|-------------------------------|--------- | Uncompressed Size | 4 GiB | | | 3.95 GiB | | Compressed Size | 3.04 GiB | | | 1.27 GiB | | **Module** | **Bandwidth
/ (MB/s)** | **Speedup** | | **Bandwidth
/ (MB/s)** | **Speedup** | gzip | 250 | 1 | | 293 | 1 | rapidgzip (0 threads) | 5060 | 20.1 | | 2070 | 6.9 | rapidgzip (1 threads) | 487 | 1.9 | | 630 | 2.1 | rapidgzip (2 threads) | 839 | 3.3 | | 694 | 2.3 | rapidgzip (6 threads) | 2365 | 9.4 | | 1740 | 5.8 | rapidgzip (12 threads) | 4116 | 16.4 | | 1900 | 6.4 | rapidgzip (24 threads) | 4974 | 19.8 | | 2040 | 6.8 | rapidgzip (32 threads) | 4612 | 18.3 | | 2580 | 8.6 ## Benchmarks for Different Compressors 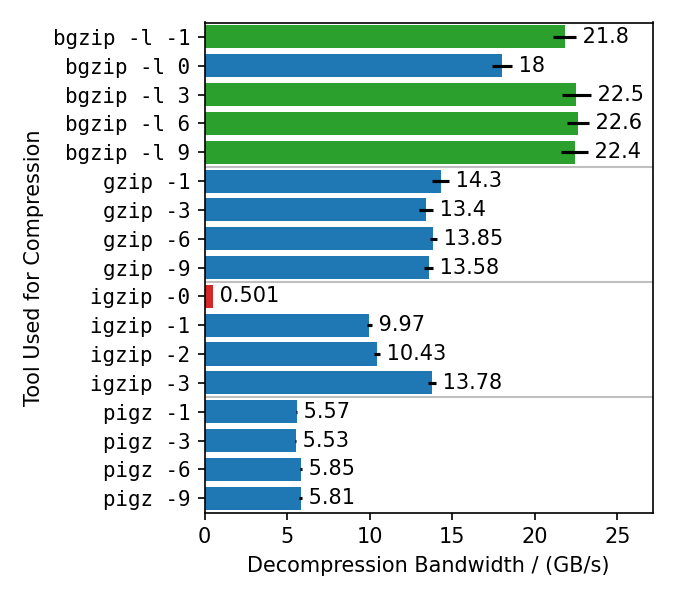 This benchmark compresses the enlarged Silesia TAR with different gzip implementations, each with different compression levels. Rapidgzip is then used to decompress the resulting files with 128 cores. Rapidgzip can parallelize decompression for almost all tested cases. The only exception is files compressed with `igzip -0`, because these files contain only a single several gigabytes large deflate block. This is the only known tool to produce such a pathological deflate block. The decompression bandwidth for the other compressors, varies quite a lot. The fastest decompression is reached with 22 GB/s for bgzip-compressed files because the bgzip format is directly supported, which enables rapidgzip to avoid the two-stage decompression method and also enables rapidgzip to offload all of the work to ISA-L. Files compressed with `bgzip -l 0` decompress slightly slower with "only" 18 GB/s, because it creates a fully non-compressed gzip stream and therefore is more I/O bound than the other bgzip-generated files. Decompression of pigz-generated files is the slowest with 6 GB/s as opposed to 10-14 GB/s for gzip and igzip. It is not clear why that is. It might be because `pigz` generates small deflate blocks and adds flush markers. The values in this chart are higher than in table 3 in the [paper](#citation) because the measurements were done with rapidgzip 0.10.1 instead of version 0.5.0. ## Benchmarks for Different Decompressors 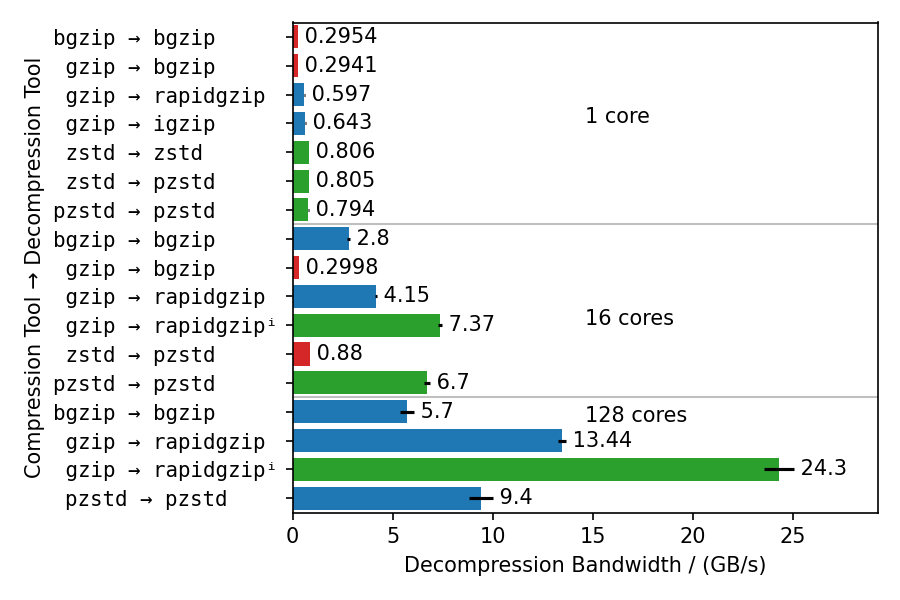 These benchmarks use different compressors and different decompressors to show multiple things: - Single-core decompression of rapidgzip is close to `igzip` and roughly twice as fast as `bgzip`, which uses zlib. - Decompression bandwidth with ISA-L can somewhat compete with zstd and is only 25% slower. - Both `bgzip` and `pzstd` can only parallelize decompression of files compressed with `bgzip` / `pzstd`. This especially means that files compressed with the standard `zstd` tool cannot be decompressed in parallel and tops out at ~800 MB/s. - Even for bgzip-compressed files, rapidgzip is always faster than `bgzip` for decompression, thanks to ISA-L and better multi-threading. - Rapidgzip scales higher than pzstd for decompression with many cores, and can be more than twice as fast when an index exists: 24.3 GB/s vs. 9.5 GB/s. The values in this chart are higher than in table 4 in the [paper](#citation) because the measurements were done with rapidgzip 0.10.1 instead of [version 0.5.0](https://raw.githubusercontent.com/mxmlnkn/indexed_bzip2/master/results/benchmarks/rapidgzip-comparison-benchmarks-2023-09-05T20-45-10/rapidgzip-0.5.0-compression-format-comparison.png).
Usage
Command Line Tool
```bash rapidgzip --help
Parallel decoding: 1.7 s
time rapidgzip -d -c -P 0 sample.gz | wc -c
Serial decoding: 22 s
time gzip -d -c sample.gz | wc -c ```
Help Output
``` A gzip decompressor tool based on the rapidgzip backend from ratarmount Usage: rapidgzip [OPTION...] positional parameters Actions options: -d, --decompress Force decompression. Only for compatibility. No compression supported anyways. --import-index arg Uses an existing gzip index. --export-index arg Write out a gzip index file. --count Prints the decompressed size. -l, --count-lines Prints the number of newline characters in the decompressed data. --analyze Print output about the internal file format structure like the block types. Advanced options: --chunk-size arg The chunk size decoded by the parallel workers in KiB. (default: 4096) --verify Verify CRC32 checksum. Will slow down decompression and there are already some implicit and explicit checks like whether the end of the file could be reached and whether the stream size is correct. --no-verify Do not verify CRC32 checksum. Might speed up decompression and there are already some implicit and explicit checks like whether the end of the file could be reached and whether the stream size is correct. --io-read-method arg Option to force a certain I/O method for reading. By default, pread will be used when possible. Possible values: pread, sequential, locked-read (default: pread) --index-format arg Option to select an output index format. Possible values: gztool, gztool-with-lines, indexed_gzip. (default: indexed_gzip) Decompression options: -c, --stdout Output to standard output. This is the default, when reading from standard input. -f, --force Force overwriting existing output files. Also forces decompression even when piped to /dev/null. -o, --output arg Output file. If none is given, use the input file name with '.gz' stripped or '.out'. If no input is read from standard input and not output file is given, then will write to standard output. -k, --keep Keep (do not delete) input file. Only for compatibility. This tool will not delete anything automatically! -P, --decoder-parallelism arg Use the parallel decoder. If an optional integer >= 1 is given, then that is the number of decoder threads to use. Note that there might be further threads being started with non-decoding work. If 0 is given, then the parallelism will be determined automatically. (default: 0) --ranges arg Decompress only the specified byte ranges. Example: 10@0,1KiB@15KiB,5L@20L to decompress the first 10 bytes, 1024 bytes at offset 15 KiB, as well as the 5 lines after skipping the first 20 lines. Output options: -h, --help Print this help message. -q, --quiet Suppress noncritical error messages. -v, --verbose Print debug output and profiling statistics. -V, --version Display software version. --oss-attributions Display open-source software licenses. --oss-attributions-yaml Display open-source software licenses in YAML format for use with Conda. If no file names are given, rapidgzip decompresses from standard input to standard output. If the output is discarded by piping to /dev/null, then the actual decoding step might be omitted if neither -l nor -L nor --force are given. Examples: Decompress a file: rapidgzip -d file.gz Decompress a file in parallel: rapidgzip -d -P 0 file.gz List information about all gzip streams and deflate blocks: rapidgzip --analyze file.gz ```Python Library
Simple open, seek, read, and close
```python3 from rapidgzip import RapidgzipFile
file = RapidgzipFile("example.gz", parallelization=os.cpu_count())
You can now use it like a normal file
file.seek(123) data = file.read(100) file.close() ```
The first call to seek will ensure that the block offset list is complete and therefore might create them first. Because of this, the first call to seek might take a while.
Use with context manager
```python3 import os import rapidgzip
with rapidgzip.open("example.gz", parallelization=os.cpu_count()) as file: file.seek(123) data = file.read(100) ```
Storing and loading the block offset map
The creation of the list of gzip blocks can take a while because it has to decode the gzip file completely. To avoid this setup when opening a gzip file, the block offset list can be exported and imported.
```python3 import os import rapidgzip
index_path = "example.gz.gzindex"
with rapidgzip.open("example.gz", parallelization=os.cpucount()) as file: file.seek(123) data = file.read(100) file.exportindex(index_path)
with rapidgzip.open("example.gz", parallelization=os.cpucount()) as file: file.importindex(index_path) file.seek(123) data = file.read(100) ```
Open a pure Python file-like object for indexed reading
```python3 import io import os import rapidgzip as rapidgzip
with open("example.gz", "rb") as file: inmemoryfile = io.BytesIO(file.read())
with rapidgzip.open(inmemoryfile, parallelization=os.cpu_count()) as file: file.seek(123) data = file.read(100) ```
Via Ratarmount
rapidgzip is the default backend in ratarmount since version 0.14.0.
Then, you can use ratarmount to mount single gzip files easily.
```bash base64 /dev/urandom | head -c $(( 4 * 1024 * 1024 * 1024 )) | gzip > sample.gz
Serial decoding: 23 s
time gzip -c -d sample.gz | wc -c
python3 -m pip install --user ratarmount ratarmount sample.gz mounted
Parallel decoding: 3.5 s
time cat mounted/sample | wc -c
Random seeking to the middle of the file and reading 1 MiB: 0.287 s
time dd if=mounted/sample bs=$(( 1024 * 1024 )) \ iflag=skipbytes,countbytes skip=$(( 2 * 1024 * 1024 * 1024 )) count=$(( 1024 * 1024 )) | wc -c ```
C++ library
Because it is written in C++, it can of course also be used as a C++ library. rapidgzip supports streaming of uncompressed data with a predefined buffer size, although small buffer sizes decrease performance.
In order to make heavy use of templates and to simplify compiling with Python setuptools, it is header-only.
The license is also permissive enough for most use cases.
Rapidgzip can be added to an existing CMake project using FetchContent, or directly via add_subdirectory
An example CMake build using FetchContent can be found here.
Citation
A paper describing the implementation details and showing the scaling behavior with up to 128 cores has been submitted to and accepted in ACM HPDC'23, The 32nd International Symposium on High-Performance Parallel and Distributed Computing. The paper can also be accessed on ACM DL or Arxiv. The accompanying presentation can be found here.
If you use this software for your scientific publication, please cite it as:
bibtex
@inproceedings{rapidgzip,
author = {Knespel, Maximilian and Brunst, Holger},
title = {Rapidgzip: Parallel Decompression and Seeking in Gzip Files Using Cache Prefetching},
year = {2023},
isbn = {9798400701559},
publisher = {Association for Computing Machinery},
address = {New York, NY, USA},
url = {https://doi.org/10.1145/3588195.3592992},
doi = {10.1145/3588195.3592992},
abstract = {Gzip is a file compression format, which is ubiquitously used. Although a multitude of gzip implementations exist, only pugz can fully utilize current multi-core processor architectures for decompression. Yet, pugz cannot decompress arbitrary gzip files. It requires the decompressed stream to only contain byte values 9–126. In this work, we present a generalization of the parallelization scheme used by pugz that can be reliably applied to arbitrary gzip-compressed data without compromising performance. We show that the requirements on the file contents posed by pugz can be dropped by implementing an architecture based on a cache and a parallelized prefetcher. This architecture can safely handle faulty decompression results, which can appear when threads start decompressing in the middle of a gzip file by using trial and error. Using 128 cores, our implementation reaches 8.7 GB/s decompression bandwidth for gzip-compressed base64-encoded data, a speedup of 55 over the single-threaded GNU gzip, and 5.6 GB/s for the Silesia corpus, a speedup of 33 over GNU gzip.},
booktitle = {Proceedings of the 32nd International Symposium on High-Performance Parallel and Distributed Computing},
pages = {295–307},
numpages = {13},
keywords = {gzip, decompression, parallel algorithm, performance, random access},
location = {Orlando, FL, USA},
series = {HPDC '23},
}
About
This tool originated as a backend for ratarmount. After writing the bzip2 backend for ratarmount, my hesitation about reimplementing custom decoders for existing file formats has vastly diminished. And, while random access to gzip files did exist with indexed_gzip, it did not support parallel decompression neither for the index creation nor when the index already exists. The latter of which is trivial, when ignoring load balancing issues, but parallelizing even the index creation is vastly more complicated because decompressing data requires the previous 32 KiB of decompressed data to be known.
After implementing a production-ready version by improving upon the algorithm used by pugz, I submitted a paper. The review process was double-blind and I was unsure whether to pseudonymize Pragzip because it has already been uploaded to Github. In the end, I used "rapidgzip" during the review process and because I was not sure, which form fields should be filled with the pseudonymized title, I simply stuck with it. Rapidgzip was chosen for similar reason to pragzip, namely the P and RA are acronyms for Parallel and Random Access. As rapgzip, did not stick, I used rapidgzip, which now also contains the foremost design goal in its name: being rapidly faster than single-threaded implementations. Furthermore, the additional ID could be interpreted to stand for Index and Decompression, making "rapid" a partial backronym.
Internal Architecture
The main part of the internal architecture used for parallelizing is the same as used for indexed_bzip2.
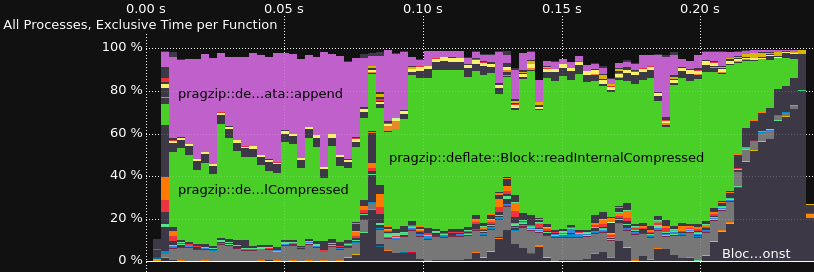
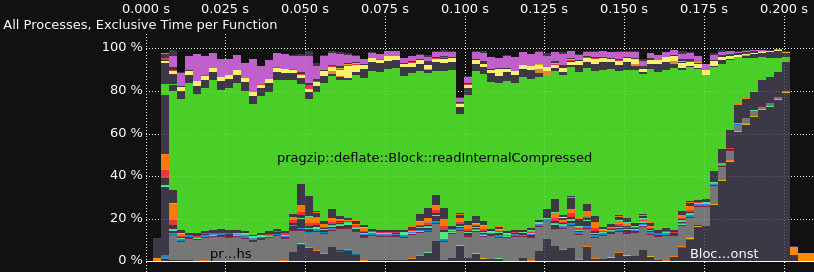
Tracing the Decoder
Performance profiling and tracing is done with Score-P for instrumentation and Vampir for visualization. This is one way, you could install Score-P with most of the functionalities on Ubuntu 22.04.
Installation of Dependencies
Installation steps for Score-P
```bash sudo apt-get install libopenmpi-dev openmpi-bin gcc-11-plugin-dev llvm-dev libclang-dev libunwind-dev \ libopen-trace-format-dev otf-trace libpapi-dev # Install Score-P (to /opt/scorep) SCOREP_VERSION=8.0 wget "https://perftools.pages.jsc.fz-juelich.de/cicd/scorep/tags/scorep-${SCOREP_VERSION}/scorep-${SCOREP_VERSION}.tar.gz" tar -xf "scorep-${SCOREP_VERSION}.tar.gz" cd "scorep-${SCOREP_VERSION}" ./configure --with-mpi=openmpi --enable-shared --without-llvm --without-shmem --without-cubelib --prefix="/opt/scorep-${SCOREP_VERSION}" make -j $( nproc ) make install # Add /opt/scorep to your path variables on shell start cat <Tracing
Results for a version from 2023-02-04
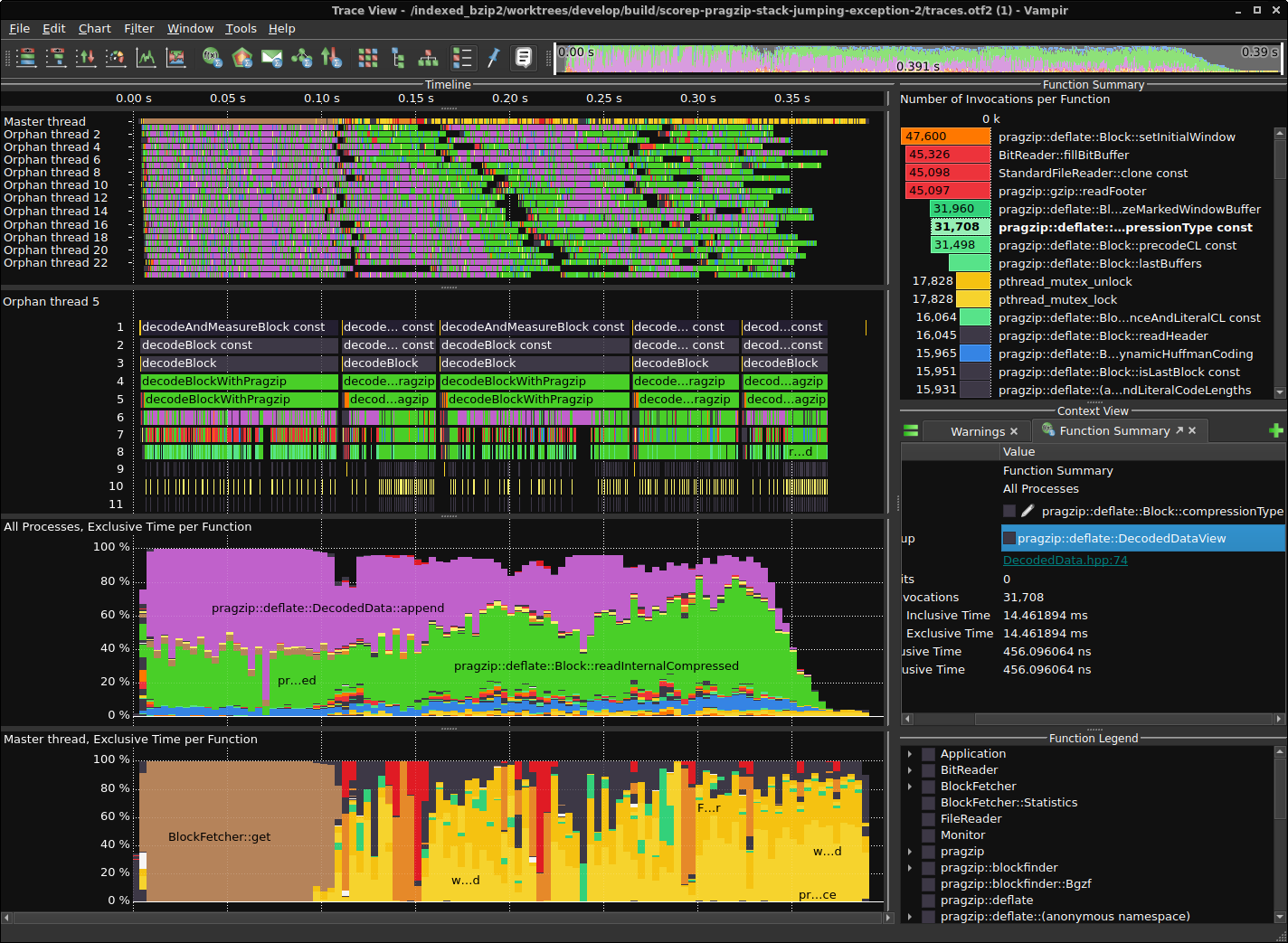
Comparison without and with rpmalloc preloaded
Owner
- Name: Maximilian Knespel
- Login: mxmlnkn
- Kind: user
- Location: Germany, Dresden
- Repositories: 38
- Profile: https://github.com/mxmlnkn
Citation (CITATION.cff)
# Validate with:
# python3 -m pip install --user cffconvert
# cffconvert --validate
cff-version: 1.2.0
title: Rapidgzip
message: >-
If you use this software, please cite it using the metadata from this file.
type: software
authors:
- given-names: Maximilian
family-names: Knespel
orcid: "https://orcid.org/0000-0001-9568-3075"
email: maximilian.knespel@tu-dresden.de
repository-code: "https://github.com/mxmlnkn/indexed_bzip2"
repository: "https://github.com/mxmlnkn/rapidgzip"
abstract: >-
A replacement for gzip for decompressing gzip files using
multiple threads.
keywords:
- Gzip
- Decompression
- Parallel Algorithm
- Performance
- Random Access
license: MIT
preferred-citation:
type: conference-paper
authors:
- given-names: Maximilian
family-names: Knespel
orcid: "https://orcid.org/0000-0001-9568-3075"
email: maximilian.knespel@tu-dresden.de
- family-names: "Brunst"
given-names: "Holger"
orcid: "https://orcid.org/0000-0003-2224-0630"
title: "Rapidgzip: Parallel Decompression and Seeking in Gzip Files Using Cache Prefetching"
year: 2023
isbn: 9798400701559
publisher:
name: "Association for Computing Machinery"
alias: "ACM"
city: "New York"
region: "NY"
country: "US"
url: "https://doi.org/10.1145/3588195.3592992"
doi: "10.1145/3588195.3592992"
abstract: >-
Gzip is a file compression format, which is ubiquitously used. Although a multitude of gzip implementations exist, only pugz can fully utilize current multi-core processor architectures for decompression. Yet, pugz cannot decompress arbitrary gzip files. It requires the decompressed stream to only contain byte values 9–126. In this work, we present a generalization of the parallelization scheme used by pugz that can be reliably applied to arbitrary gzip-compressed data without compromising performance. We show that the requirements on the file contents posed by pugz can be dropped by implementing an architecture based on a cache and a parallelized prefetcher. This architecture can safely handle faulty decompression results, which can appear when threads start decompressing in the middle of a gzip file by using trial and error. Using 128 cores, our implementation reaches 8.7 GB/s decompression bandwidth for gzip-compressed base64-encoded data, a speedup of 55 over the single-threaded GNU gzip, and 5.6 GB/s for the Silesia corpus, a speedup of 33 over GNU gzip.
collection-title: "Proceedings of the 32nd International Symposium on High-Performance Parallel and Distributed Computing"
start: 295
end: 307
pages: 13
keywords:
- Gzip
- Decompression
- Parallel Algorithm
- Performance
- Random Access
conference:
name: "The 32nd International Symposium on High-Performance Parallel and Distributed Computing"
alias: "HPDC '23"
city: "Orlando"
date-start: 2023-06-20
date-end: 2023-06-23
region: "FL"
country: "US"
website: "https://www.hpdc.org/2023"
date-published: 2023-08-07
status: advance-online
GitHub Events
Total
- Create event: 8
- Release event: 2
- Issues event: 22
- Watch event: 56
- Delete event: 4
- Issue comment event: 50
- Push event: 11
- Pull request review event: 10
- Pull request review comment event: 12
- Pull request event: 7
- Fork event: 5
Last Year
- Create event: 8
- Release event: 2
- Issues event: 22
- Watch event: 56
- Delete event: 4
- Issue comment event: 50
- Push event: 11
- Pull request review event: 10
- Pull request review comment event: 12
- Pull request event: 7
- Fork event: 5
Committers
Last synced: 11 months ago
Committer Domains (Top 20 + Academic)
Issues and Pull Requests
Last synced: 6 months ago
All Time
- Total issues: 56
- Total pull requests: 6
- Average time to close issues: 2 months
- Average time to close pull requests: 8 days
- Total issue authors: 22
- Total pull request authors: 3
- Average comments per issue: 3.61
- Average comments per pull request: 0.67
- Merged pull requests: 4
- Bot issues: 0
- Bot pull requests: 0
Past Year
- Issues: 16
- Pull requests: 5
- Average time to close issues: 30 days
- Average time to close pull requests: 10 days
- Issue authors: 10
- Pull request authors: 2
- Average comments per issue: 2.44
- Average comments per pull request: 0.8
- Merged pull requests: 4
- Bot issues: 0
- Bot pull requests: 0
Top Authors
Issue Authors
- mxmlnkn (17)
- pdjstone (6)
- AndreiCherniaev (5)
- koriege (5)
- Vadiml1024 (3)
- darked89 (2)
- biocodz (2)
- tooptoop4 (2)
- agoertz-fls (1)
- asangphukieo (1)
- osevan (1)
- cal-pratt (1)
- moizarafat (1)
- sergeevabc (1)
- amir-sabbaghi (1)
Pull Request Authors
- cal-pratt (4)
- mxmlnkn (2)
- amir-sabbaghi (1)
Top Labels
Issue Labels
Pull Request Labels
Packages
- Total packages: 1
-
Total downloads:
- pypi 17,886 last-month
- Total dependent packages: 2
- Total dependent repositories: 0
- Total versions: 30
- Total maintainers: 1
pypi.org: rapidgzip
Parallel random access to gzip files
- Homepage: https://github.com/mxmlnkn/rapidgzip
- Documentation: https://rapidgzip.readthedocs.io/
- License: MIT
-
Latest release: 0.15.2
published 6 months ago
Rankings
Maintainers (1)
Dependencies
- actions/checkout v3 composite
- github/codeql-action/analyze v2 composite
- github/codeql-action/init v2 composite
- actions/checkout v3 composite
- conda-incubator/setup-miniconda v2 composite
- actions/checkout v3 composite
- actions/setup-python v4 composite
- conda-incubator/setup-miniconda v2 composite
- actions/checkout v3 composite
- actions/checkout v3 composite
- actions/setup-python v4 composite
- actions/checkout v3 composite
- actions/setup-python v4 composite
- actions/checkout v3 composite
- conda-incubator/setup-miniconda v2 composite
- actions/checkout v3 composite
- actions/setup-python v4 composite
- conda-incubator/setup-miniconda v2 composite
- actions/checkout v3 composite
- actions/setup-python v4 composite
- actions/upload-artifact v3 composite