Science Score: 77.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
✓CITATION.cff file
Found CITATION.cff file -
✓codemeta.json file
Found codemeta.json file -
✓.zenodo.json file
Found .zenodo.json file -
✓DOI references
Found 11 DOI reference(s) in README -
✓Academic publication links
Links to: zenodo.org -
✓Committers with academic emails
1 of 9 committers (11.1%) from academic institutions -
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (14.1%) to scientific vocabulary
Keywords
Keywords from Contributors
Repository
Create "Table 1" for research papers in Python
Basic Info
- Host: GitHub
- Owner: tompollard
- License: mit
- Language: Python
- Default Branch: main
- Homepage: https://pypi.python.org/pypi/tableone/
- Size: 1.73 MB
Statistics
- Stars: 183
- Watchers: 9
- Forks: 44
- Open Issues: 28
- Releases: 36
Topics
Metadata Files
README.md
tableone
tableone is a package for creating "Table 1" summary statistics for a patient population. It was inspired by the R package of the same name by Yoshida and Bohn.
Suggested citation
If you use tableone in your study, please cite the following paper:
Tom J Pollard, Alistair E W Johnson, Jesse D Raffa, Roger G Mark; tableone: An open source Python package for producing summary statistics for research papers, JAMIA Open, https://doi.org/10.1093/jamiaopen/ooy012
Documentation
For documentation, see: http://tableone.readthedocs.io/en/latest/. An executable demonstration of the package is available on GitHub as a Jupyter Notebook. The easiest way to try out this notebook is to open it in Google Colaboratory. A paper describing our motivations for creating the package is available at: https://doi.org/10.1093/jamiaopen/ooy012.
A note for users of tableone
While we have tried to use best practices in creating this package, automation of even basic statistical tasks can be unsound if done without supervision. We encourage use of tableone alongside other methods of descriptive statistics and, in particular, visualization to ensure appropriate data handling.
It is beyond the scope of our documentation to provide detailed guidance on summary statistics, but as a primer we provide some considerations for choosing parameters when creating a summary table at: http://tableone.readthedocs.io/en/latest/bestpractice.html.
Guidance should be sought from a statistician when using tableone for a research study, especially prior to submitting the study for publication.
Overview
At a high level, you can use the package as follows:
- Import the data into a pandas DataFrame
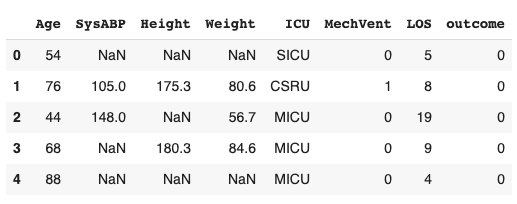
- Run tableone on this dataframe to output summary statistics
- Specify your desired output format: text, latex, markdown, etc.
Additional options include:
- Select a subset of columns.
- Specify the data type (e.g.
categorical,numerical,nonnormal). - Compute p-values, and adjust for multiple testing (e.g. with the Bonferroni correction).
- Compute standardized mean differences (SMDs).
- Provide a list of alternative labels for variables
- Limit the output of categorical variables to the top N rows.
- Display remarks relating to the appopriateness of summary measures (for example, computing tests for multimodality and normality).
Installation
To install the package with pip, run:
pip install tableone
To install this package with conda, run:
conda install -c conda-forge tableone
Example usage
- Import libraries:
python
from tableone import TableOne, load_dataset
import pandas as pd
- Load sample data into a pandas dataframe:
python
data=load_dataset('pn2012')
- Optionally, a list of columns to be included in Table 1:
python
columns = ['Age', 'SysABP', 'Height', 'Weight', 'ICU', 'death']
- Optionally, a list of columns containing categorical variables:
python
categorical = ['ICU', 'death']
- Optionally, a list of columns containing continuous variables:
python
continuous = ['Age', 'SysABP', 'Height', 'Weight']
- Optionally, a categorical variable for stratification, a list of non-normal variables, and a dictionary of alternative labels:
python
groupby = 'death'
nonnormal = ['Age']
rename={'death': 'mortality'}
- Create an instance of TableOne with the input arguments:
python
mytable = TableOne(data, columns=columns, categorical=categorical, continuous=continuous, groupby=groupby, nonnormal=nonnormal, rename=rename, pval=False)
- Display the table using the
tabulatemethod. Thetablefmtargument allows the table to be displayed in multiple formats, including "github", "grid", "fancy_grid", "rst", "html", and "latex".
python
print(mytable.tabulate(tablefmt = "fancy_grid"))
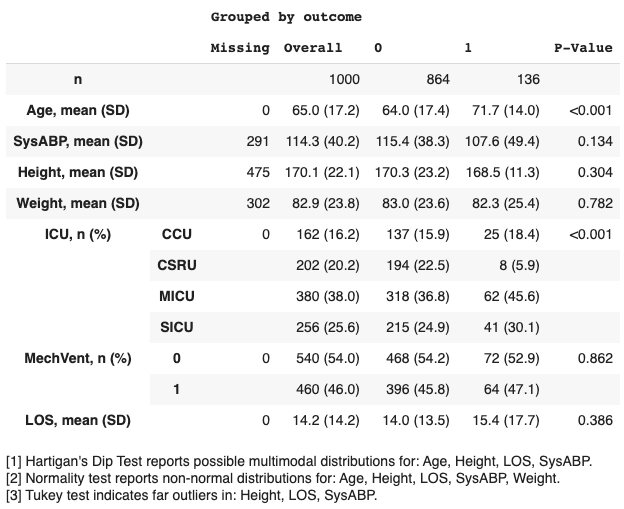
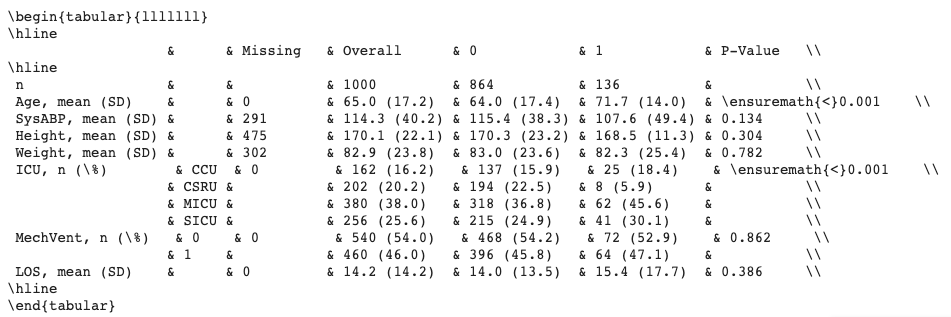
- ...which prints the following table to screen:
Grouped by mortality:
| | | Missing | 0 | 1 |
| --------- | ------ | -------- | -------------- | -------------- |
| n | | | 864 | 136 |
| Age | | 0 | 66 [52,78] | 75 [62,83] |
| SysABP | | 291 | 115.36 (38.34) | 107.57 (49.43) |
| Height | | 475 | 170.33 (23.22) | 168.51 (11.31) |
| Weight | | 302 | 83.04 (23.58) | 82.29 (25.40) |
| ICU | CCU | 0 | 137 (15.86) | 25 (18.38) |
| | CSRU | | 194 (22.45) | 8 (5.88) |
| | MICU | | 318 (36.81) | 62 (45.59) |
| | SICU | | 215 (24.88) | 41 (30.15) |
| mortality | 0 | 0 | 864 (100.0) | |
| | 1 | | | 136 (100.0) |
- Tables can be exported to file in various formats, including LaTeX, CSV, and HTML. Files are exported by calling the
to_formatmethod on the tableone object. For example, mytable can be exported to an Excel spreadsheet named 'mytable.xlsx' with the following command:
python
mytable.to_excel('mytable.xlsx')
Owner
- Name: Tom Pollard
- Login: tompollard
- Kind: user
- Location: Cambridge, MA
- Company: MIT
- Twitter: tompollard
- Repositories: 91
- Profile: https://github.com/tompollard
Citation (CITATION.cff)
cff-version: 1.2.0 message: "If you use this software, please cite it as below." authors: - family-names: "Pollard" given-names: "Tom" orcid: "https://orcid.org/0000-0002-5676-7898" - family-names: "Johnson" given-names: "Alistair" orcid: "https://orcid.org/0000-0002-8735-3014" - family-names: "Raffa" given-names: "Jesse" orcid: "https://orcid.org/0000-0001-8665-2429" - family-names: "Mark" given-names: "Roger" orcid: "https://orcid.org/0000-0002-6318-2978" title: "tableone: An open source Python package for producing summary statistics for research papers" doi: 10.1093/jamiaopen/ooy012 date-released: 2018-05-23 url: "https://doi.org/10.1093/jamiaopen/ooy012"
GitHub Events
Total
- Create event: 13
- Release event: 3
- Issues event: 12
- Watch event: 23
- Delete event: 12
- Issue comment event: 21
- Push event: 16
- Pull request review event: 2
- Pull request event: 26
- Fork event: 6
Last Year
- Create event: 13
- Release event: 3
- Issues event: 12
- Watch event: 23
- Delete event: 12
- Issue comment event: 21
- Push event: 16
- Pull request review event: 2
- Pull request event: 26
- Fork event: 6
Committers
Last synced: 9 months ago
Top Committers
| Name | Commits | |
|---|---|---|
| Tom Pollard | t****d@m****u | 444 |
| Alistair Johnson | a****j@g****m | 48 |
| Lucas Bulgarelli | l****i@e****m | 11 |
| Jesse Raffa | j****a | 7 |
| dependabot[bot] | 4****] | 4 |
| Alexander Seiler | s****x@g****m | 2 |
| jtleider | j****r@g****m | 1 |
| Ryo Fujimori | f****3@g****m | 1 |
| Vincent Lequertier | v****e@a****g | 1 |
Committer Domains (Top 20 + Academic)
Issues and Pull Requests
Last synced: 6 months ago
All Time
- Total issues: 74
- Total pull requests: 84
- Average time to close issues: 7 months
- Average time to close pull requests: 16 days
- Total issue authors: 47
- Total pull request authors: 12
- Average comments per issue: 2.85
- Average comments per pull request: 0.46
- Merged pull requests: 73
- Bot issues: 0
- Bot pull requests: 7
Past Year
- Issues: 8
- Pull requests: 28
- Average time to close issues: about 16 hours
- Average time to close pull requests: 2 days
- Issue authors: 6
- Pull request authors: 3
- Average comments per issue: 0.75
- Average comments per pull request: 0.07
- Merged pull requests: 25
- Bot issues: 0
- Bot pull requests: 4
Top Authors
Issue Authors
- tompollard (22)
- JohannesWiesner (3)
- welyt (2)
- gothmania (2)
- shahzeb1 (2)
- eroell (2)
- inescgu (1)
- Brandon96-lab (1)
- Ryo-F (1)
- proc1125 (1)
- lgracen (1)
- rherman9 (1)
- rutgervandeleur (1)
- adhaimovich (1)
- christian-geier (1)
Pull Request Authors
- tompollard (72)
- dependabot[bot] (9)
- lbulgarelli (3)
- cipherome-minkim (2)
- goggle (2)
- jakob1379 (2)
- jowinjestine (2)
- alistairewj (2)
- jraffa (2)
- jtleider (1)
- Ryo-F (1)
- 260147169 (1)
Top Labels
Issue Labels
Pull Request Labels
Packages
- Total packages: 2
-
Total downloads:
- pypi 30,842 last-month
-
Total dependent packages: 4
(may contain duplicates) -
Total dependent repositories: 33
(may contain duplicates) - Total versions: 111
- Total maintainers: 3
pypi.org: tableone
tableone is a package for creating 'Table 1' summary statistics for a patient population.
- Documentation: https://tableone.readthedocs.io/
- License: MIT License
-
Latest release: 0.9.5
published 10 months ago
Rankings
Maintainers (3)
conda-forge.org: tableone
tableone is a package for creating Table 1 for research papers in Python.
- Homepage: http://github.com/tompollard/tableone
- License: MIT
-
Latest release: 0.7.12
published over 3 years ago
Rankings
Dependencies
- nose *
- numpy ==1.12.1
- pandas ==0.22.0
- scipy ==0.18.1
- statsmodels ==0.8.0
- tabulate ==0.8.2
- numpy >=1.12.1
- pandas >=0.22.0
- scipy >=0.18.1
- statsmodels >=0.8.0
- tabulate >=0.8.2
- actions/checkout v3 composite
- actions/setup-python v3 composite
- actions/upload-artifact v3 composite
- Jinja2 *
- numpy *
- openpyxl *
- pandas *
- scipy *
- statsmodels *
- tabulate *