ptmvision
Web application for the interactive visualization and exploration of post-translational modifications of proteins from open and closed search software.
Science Score: 57.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
✓CITATION.cff file
Found CITATION.cff file -
✓codemeta.json file
Found codemeta.json file -
✓.zenodo.json file
Found .zenodo.json file -
✓DOI references
Found 1 DOI reference(s) in README -
○Academic publication links
-
○Academic email domains
-
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (9.9%) to scientific vocabulary
Keywords
Repository
Web application for the interactive visualization and exploration of post-translational modifications of proteins from open and closed search software.
Basic Info
- Host: GitHub
- Owner: Integrative-Transcriptomics
- License: gpl-3.0
- Language: HTML
- Default Branch: main
- Homepage: https://ptmvision-tuevis.cs.uni-tuebingen.de/
- Size: 242 MB
Statistics
- Stars: 4
- Watchers: 4
- Forks: 0
- Open Issues: 0
- Releases: 0
Topics
Metadata Files
README.md

PTMVision
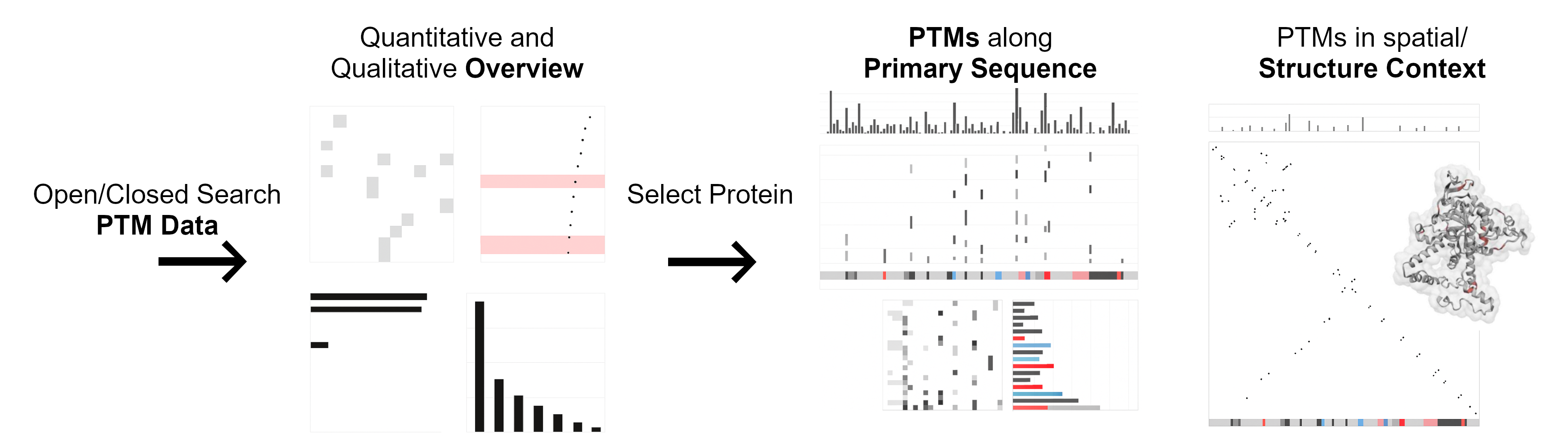
PTMVision is an interactive web-based platform for the visual exploration of post-translational modifications (PTMs) identified in mass spectrometry-based proteomics data. It enables researchers to interpret the complex PTM landscape, supporting insights into molecular mechanisms of biological processes and disease.
🔗 Web server: https://ptmvision-tuevis.cs.uni-tuebingen.de
✨ Features
- Rich interactive visualizations for sample-level and protein-level PTM data
- Linked plots for seamless navigation and exploration
- 3D structure integration and contact map analysis for structural context
- Highlights residue-residue PTM interactions in close spatial proximity
- Annotation integration from UniProt and UniMod for interpretability
- Support for multiple search engine output formats and a custom CSV format
- Session export/import for reproducibility and collaboration
- Intuitive UI accessible to users without programming expertise
📁 Supported Input Formats
PTMVision accepts output from several widely-used search engines:
| Search Engine | Tested Versions | Key Columns | Processing Steps |
| ------------------------------------------ | ---------------------------------- | ------------------------------------------------------------------------------------------- | ----------------------------------------------------------------------------------------------------------------------------- |
| ionbot | v0.10.0, v0.11.0 | uniprot_id, unexpected_modification, position | Direct parsing of modification information |
| MSFragger (via FragPipe + PTMShepherd) | MSFragger v4.0, PTMShepherd v2.0.6 | Modified Peptide, Assigned Modifications, Localization, Protein ID, Protein Start | Filtering ambiguous or multi-localized sites; mass shift mapping to UniMod; UniProt sequence alignment; UniMod classification |
| Sage | v0.13.1 | See psm-utils | FDR filtering; decoy removal; UniMod mapping; PTM localization from variable mods only |
| MaxQuant | v2.4.13.0 | Search result columns used for mapping | Mapping MaxQuant-specific names to UniMod; resolving multi-mapping peptides; UniProt mapping |
| Spectronaut (PTM Site Report) | v9 | PTM.ProteinId, PTM.SiteLocation, PTM.ModificationTitle, PTM.SiteAA | Infers mass shifts and UniMod classifications from reported data and UniProt sequence |
Note: For FragPipe-derived data, localization confidence is inferred from the
MSFragger Localizationfield. Use caution until future versions provide dedicated localization scores.
📄 Custom Plain CSV Format
Users can submit manually compiled PTM data using a structured CSV format when other formats are unavailable.
Required Columns
| Column Name | Description |
| ----------------------------- | ------------------------------------------------------------------------ |
| uniprot_id | UniProt accession of the protein |
| position | 1-based index of the modified residue |
| modification_unimod_name | Name of the UniMod modification |
| modification_unimod_id | UniMod ID |
| mass_shift (optional) | Monoisotopic mass shift in Daltons (inferred from UniMod ID if omitted) |
| classification (optional) | UniMod classification (inferred from ID and sequence context if omitted) |
Example entry:
P04075,5,carbamidomethyl,4,57.021464,Artefact
⚠ Note: Classification relies on up-to-date UniProt sequences. Sequence version mismatches may lead to misaligned positions.
🐳 Running Locally via Docker
To deploy PTMVision locally:
```bash
Clone repository
git clone https://github.com/Integrative-Transcriptomics/PTMVision.git cd PTMVision
Build Docker image
docker build --tag ptmvision app
Run locally
docker run -dp 127.0.0.1:5001:5001 ptmvision ```
Note: Prebuilt Docker images are not currently provided.
🙋 Need Help?
- 📖 Read the Usage Guide
- 🐛 Found an issue or missing format? Open a GitHub issue
✏️ Citation
If you use this software, please cite it as below:
``` PTMVision: An Interactive Visualization Webserver for Post-translational Modifications of Proteins
Simon Hackl, Caroline Jachmann, Mathias Witte Paz, Theresa Anisja Harbig, Lennart Martens, and Kay Nieselt
Journal of Proteome Research 2025 24 (2), 919-928
DOI: 10.1021/acs.jproteome.4c00679 ```
Owner
- Name: Integrative-Transcriptomics
- Login: Integrative-Transcriptomics
- Kind: organization
- Repositories: 15
- Profile: https://github.com/Integrative-Transcriptomics
Citation (CITATION.cff)
# This CITATION.cff file was generated with cffinit.
# Visit https://bit.ly/cffinit to generate yours today!
cff-version: 1.2.0
title: >-
PTMVision: An Interactive Visualization Webserver for
Post-translational Modifications of Proteins
message: >-
If you use this software, please cite it using the
metadata from this file.
type: software
authors:
- given-names: Simon
family-names: Hackl
orcid: "https://orcid.org/0009-0000-3596-3411"
- given-names: Caroline
family-names: Jachmann
orcid: "https://orcid.org/0009-0000-4679-7295"
- given-names: Mathias
family-names: Witte Paz
orcid: "https://orcid.org/0000-0001-8113-4139"
- given-names: Theresa Anisja
family-names: Harbig
orcid: "https://orcid.org/0000-0001-8716-5788"
- given-names: Lennart
family-names: Martens
orcid: "https://orcid.org/0000-0003-4277-658X"
- given-names: Kay
family-names: Nieselt
orcid: "https://orcid.org/0000-0002-1283-7065"
identifiers:
- type: doi
value: 10.1021/acs.jproteome.4c00679
description: Article
- type: other
value: "39772617"
description: PMID
repository-code: "https://github.com/Integrative-Transcriptomics/PTMVision"
url: "https://ptmvision-tuevis.cs.uni-tuebingen.de/"
repository: "https://doi.org/10.5281/zenodo.13270568"
abstract: >-
Recent improvements in methods and instruments used in
mass spectrometry have greatly enhanced the detection of
protein post-translational modifications (PTMs). On the
computational side, the adoption of open modification
search strategies now allows for the identification of a
wide variety of PTMs, potentially revealing hundreds to
thousands of distinct modifications in biological samples.
While the observable part of the proteome is continuously
growing, the visualization and interpretation of this vast
amount of data in a comprehensive fashion is not yet
possible. There is a clear need for methods to easily
investigate the PTM landscape and to thoroughly examine
modifications on proteins of interest from acquired mass
spectrometry data. We present PTMVision, a web server
providing an intuitive and simple way to interactively
explore PTMs identified in mass spectrometry-based
proteomics experiments and to analyze the modification
sites of proteins within relevant context. It offers a
variety of tools to visualize the PTM landscape from
different angles and at different levels, such as 3D
structures and contact maps, UniMod classification
summaries, and site specific overviews. The web server’s
user-friendly interface ensures accessibility across
diverse scientific backgrounds.
keywords:
- post-translational modifications
- open search
- visualization
- Web server
license: GPL-3.0
commit: 117dfdfea1a600f374e4bc85bef02dd57959920a
version: "2.0"
date-released: "2025-01-08"
preferred-citation:
type: article
authors:
- given-names: Simon
family-names: Hackl
orcid: "https://orcid.org/0009-0000-3596-3411"
- given-names: Caroline
family-names: Jachmann
orcid: "https://orcid.org/0009-0000-4679-7295"
- given-names: Mathias
family-names: Witte Paz
orcid: "https://orcid.org/0000-0001-8113-4139"
- given-names: Theresa Anisja
family-names: Harbig
orcid: "https://orcid.org/0000-0001-8716-5788"
- given-names: Lennart
family-names: Martens
orcid: "https://orcid.org/0000-0003-4277-658X"
- given-names: Kay
family-names: Nieselt
orcid: "https://orcid.org/0000-0002-1283-7065"
doi: "10.1021/acs.jproteome.4c00679"
journal: "Journal of Proteome Research"
start: 919 # First page number
end: 928 # Last page number
title: "PTMVision: An Interactive Visualization Webserver for Post-translational Modifications of Proteins"
volume: 24
number: 2
year: 2025
note: "PMID: 39772617"
URL: "https://doi.org/10.1021/acs.jproteome.4c00679"
eprint: "https://doi.org/10.1021/acs.jproteome.4c00679"
GitHub Events
Total
- Watch event: 3
- Delete event: 3
- Push event: 33
- Pull request event: 2
- Fork event: 1
- Create event: 1
Last Year
- Watch event: 3
- Delete event: 3
- Push event: 33
- Pull request event: 2
- Fork event: 1
- Create event: 1
Dependencies
- python 3.8-slim-buster build
- Bio ==1.6.2
- Flask ==2.2.3
- Requests ==2.31.0
- Werkzeug ==2.2.2
- biopython ==1.81
- brotlipy ==0.7.0
- cachelib ==0.12.0
- flask_session ==0.7.0
- gunicorn ==21.2.0
- numpy ==1.24.4
- pandas ==1.5.3
- psm_utils ==0.7.4
- pyteomics ==4.7.1
- python-dotenv ==1.0.1
- scipy ==1.10.0
- setuptools ==68.0.0
- unimod_mapper ==0.6.6
- Flask-Session *
- biopython *
- brotli *
- datetime *
- flask *
- gunicorn *
- numpy *
- psm_utils *
- python-dotenv *
- scipy *
- unimod-mapper *