gitprojecthealth
A Mining Software Repository and Ticket management Platform tool, in Pharo
Science Score: 67.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
✓CITATION.cff file
Found CITATION.cff file -
✓codemeta.json file
Found codemeta.json file -
✓.zenodo.json file
Found .zenodo.json file -
✓DOI references
Found 2 DOI reference(s) in README -
✓Academic publication links
Links to: zenodo.org -
○Academic email domains
-
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (10.3%) to scientific vocabulary
Keywords
Repository
A Mining Software Repository and Ticket management Platform tool, in Pharo
Basic Info
- Host: GitHub
- Owner: moosetechnology
- Language: Smalltalk
- Default Branch: main
- Homepage: https://modularmoose.org/users/git-project-health/getting-started-with-gitproject-health/
- Size: 1.87 MB
Statistics
- Stars: 3
- Watchers: 10
- Forks: 3
- Open Issues: 4
- Releases: 6
Topics
Metadata Files
README.md
GitProject health
This project includes a model, an importer, and some visulization to evaluate the health of a GitLab or GitHub group.
Installation
Download a Moose image.
In the Moose image, in a playground (Ctrl+O, Ctrl+W), perform:
st
Metacello new
repository: 'github://moosetechnology/GitProjectHealth:main/src';
baseline: 'GitLabHealth';
onConflict: [ :ex | ex useLoaded ];
onUpgrade: [ :ex | ex useIncoming ];
onDowngrade: [ :ex | ex useLoaded ];
load
Usages
Import
Group import: GitLab
In a playground (Ctrl+O, Ctrl+W).
```st glhModel := GLHModel new.
glhApi := GLHApi new
privateToken: '
glhImporter := GLHModelImporter new glhApi: glhApi; glhModel: glhModel.
"137 is the ID of the a Group, you can find the number in the webpage of every project and group" glhImporter importGroup: 137. ```
Group import: GitHub
In a playground (Ctrl+O, Ctrl+W).
```st glhModel := GLHModel new.
githubImporter := GHModelImporter new glhModel: glhModel; privateToken: '
githubImporter importGroup: 'moosetechnology'. ```
More commits extracted
GitLab API only
You might want to gather more commits for a specific repository. To do so in GitLab, we added the following API
```st
myProject := ((glhModel allWithType: GLHProject) select: [ :project | project name = '
glhImporter importCommitsOf: myProject withStats: true until: '2023-01-01' asDate. ```
Visualize
To visualize the group "health"
st
dritGroup := (glhModel allWithType: GLHGroup) detect: [ :group | group id = 137 ].
canvas := (GLHGroupVisualization new forGroup: dritGroup).
canvas open.
Export
To export the visualization as a svg image
```st dritGroup := (glhModel allWithType: GLHGroup) detect: [ :group | group id = 137 ]. canvas := (GLHGroupVisualization new forGroup: dritGroup). canvas open.
canvas svgExporter withoutFixedShapes; fileName: 'drit-group-health'; export. ```
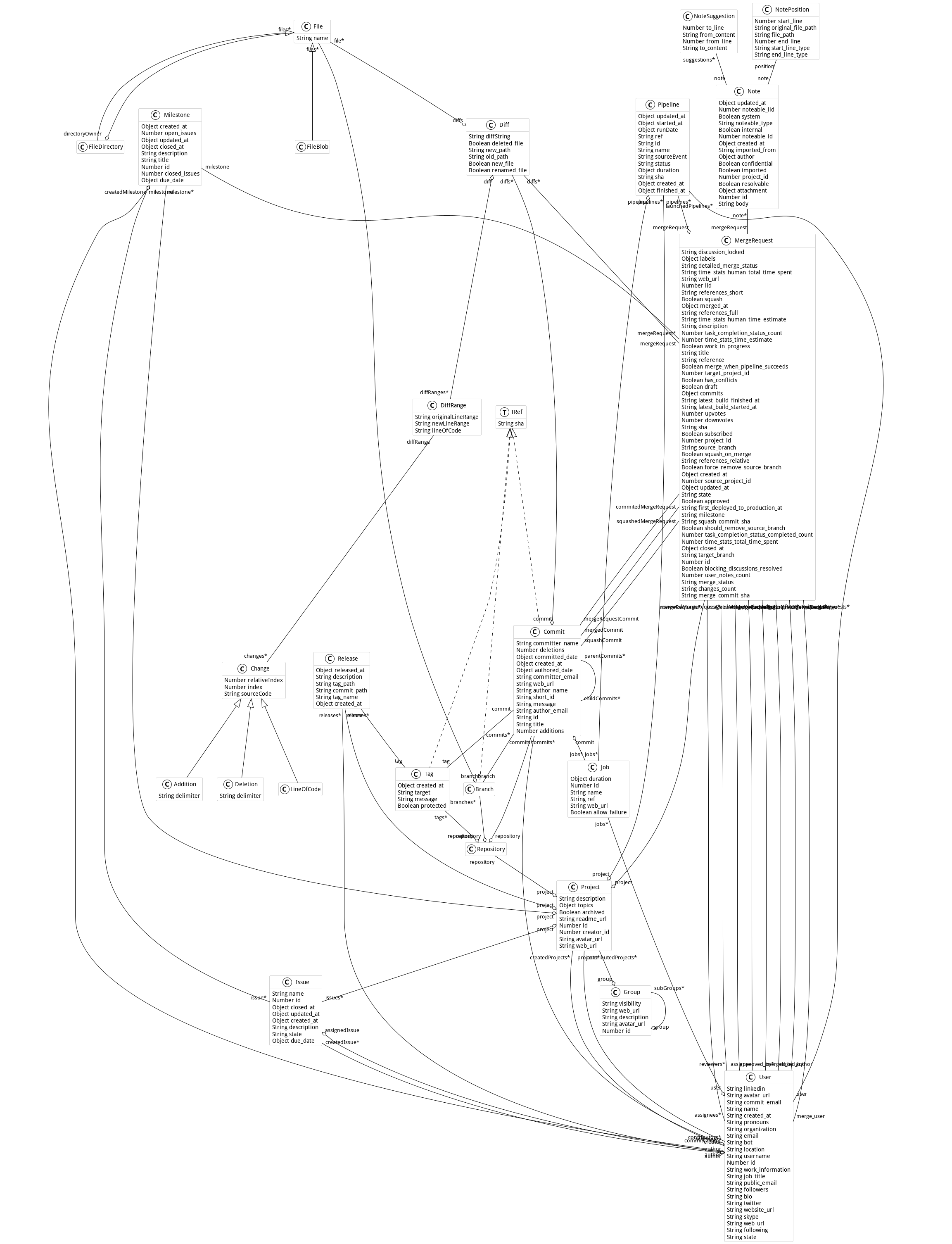
Metamodel
Here is the metamodel used in this project
Connectors
This project comes with connectors to other metamodels to increase its powerfulness. Explore this part of the documentation on the main website.
Contributor
This work has been first developed by the research department of Berger-Levrault
Running metrics with docker
Running locally
```smalltalk
|glphModel glphApi glhImporter beforeExp duringExp usersWithProjects gme|
"This example set up and run a GitProjectHealth metrics over two period of time of a given set of users and their projects. It ouputs a csv files containing : churn code, commits frequencies, code addition and deletion, comments added (e.g. // # /**/ ), avg delay before first churn and merge request duration. "
"load githealth project into your image" Metacello new repository: 'github://moosetechnology/GitProjectHealth:GLPH-importer-new-changes/src'; baseline: 'GitLabHealth'; onConflict: [ :ex | ex useIncoming ]; onUpgrade: [ :ex | ex useIncoming ]; onDowngrade: [ :ex | ex useLoaded ]; ignoreImage; load.
"set up a log at your root" TinyLogger default addFileLoggerNamed: 'pharo-code-churn.log'.
"new model instance" glphModel := GLPHEModel new.
"new API class instance"
glphApi := GLPHApi new
privateToken: '
"new importer instance" glhImporter := GLPHModelImporter new glhApi: glphApi; glhModel: glphModel; withFiles: false; withCommitDiffs: true.
"setting up the period to compare (e.g. before a experience and during an experience)" beforeExp := { #since -> ('1 march 2023' asDate). #until -> ('24 may 2023' asDate). } asDictionary . duringExp := { #since -> ('1 march 2024' asDate). #until -> ('24 may 2024' asDate). } asDictionary .
usersWithProjects := { " 'dev nameA' -> { projectID1 . projectID2 }." " 'dev nameB' -> { projectID3 . projectID2 }." 'John Do' -> { 14 . 543 . 2455 }. } asDictionary.
gme := GitMetricExporter new glhImporter: glhImporter; initEntitiesFromUserProjects: usersWithProjects; beforeDic: beforeExp; duringDic: duringExp; label: 'GitLabHealth'.
"select among the following calendar class (at least one) " gme exportOver: { Date . Week . Month . Year .}.
"the output files are located at 'FileLocator home/*.csv' " Smalltalk snapshot: true andQuit: true. ```
deploying with docker
```bash git clone https://github.com/moosetechnology/GitProjectHealth.git cd GitProjectHealth git checkout GLPH-importer-new-changes
sudo docker build -t code-churn-pharo . sudo docker run code-churn-pharo & ```
Locate and retrieve csv output files:
bash
sudo docker ps
sudo docker exec -it <container-id> find / -type f -name 'IA4Code*.csv' 2>/dev/null
Owner
- Name: moosetechnology
- Login: moosetechnology
- Kind: organization
- Website: https://modularmoose.org/
- Repositories: 28
- Profile: https://github.com/moosetechnology
Citation (CITATION.cff)
cff-version: 1.2.0 message: "If you use this software, please cite it as below." authors: - family-names: "Verhaeghe" given-names: "Benoit" orcid: "https://orcid.org/0000-0002-4588-2698" - family-names: "Hlad" given-names: "Nicolas" orcid: "https://orcid.org/0000-0003-4989-2508" - family-names: "Bauvent" given-names: "Kilian" - family-names: "Anquetil" given-names: "Nicolas" orcid: "https://orcid.org/0000-0003-1486-8399" title: "GitProjectHealth" version: v1.1.0 doi: 10.5281/zenodo.13886504 date-released: 2024-10-24 url: "https://github.com/moosetechnology/GitProjectHealth"
GitHub Events
Total
- Create event: 97
- Release event: 5
- Issues event: 7
- Watch event: 1
- Delete event: 105
- Member event: 2
- Issue comment event: 20
- Push event: 324
- Pull request review comment event: 100
- Pull request review event: 148
- Pull request event: 234
- Fork event: 3
Last Year
- Create event: 97
- Release event: 5
- Issues event: 7
- Watch event: 1
- Delete event: 105
- Member event: 2
- Issue comment event: 20
- Push event: 324
- Pull request review comment event: 100
- Pull request review event: 148
- Pull request event: 234
- Fork event: 3