pretzel
Javascript full-stack framework for Big Data visualisation and analysis
Science Score: 67.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
✓CITATION.cff file
Found CITATION.cff file -
✓codemeta.json file
Found codemeta.json file -
✓.zenodo.json file
Found .zenodo.json file -
✓DOI references
Found 6 DOI reference(s) in README -
○Academic publication links
-
✓Committers with academic emails
8 of 21 committers (38.1%) from academic institutions -
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (14.2%) to scientific vocabulary
Keywords
Keywords from Contributors
Repository
Javascript full-stack framework for Big Data visualisation and analysis
Basic Info
Statistics
- Stars: 45
- Watchers: 6
- Forks: 13
- Open Issues: 155
- Releases: 28
Topics
Metadata Files
README.md

This README gives a high level overview of what Pretzel can do. More detailed documentation is being developed at https://docs.plantinformatics.io/.
What is Pretzel
Pretzel is a web-based online framework for the real-time interactive display integration of genetic and genomic datasets. It is built on Ember.js (front end), Loopback.js (back end) and D3.js (visualisation).
Get started now
Agriculture Victoria hosts the following Pretzel instances for the following species for public datasets:
| Species | URL | |--|--| | Wheat | https://plantinformatics.io/ | | Pulses | https://pulses.plantinformatics.io/ | | Barley | https://barley.plantinformatics.io/ |
Users can sign up for an account by following the Sign Up links in the top right of the page.
Pretzel features
Integration of a diverse range of genetic and genomic information
Genetic map and chromosome-scale genomic assembly alignments
Visualisation of genomic features, including genes, markers and QTLs
Genotype data visualisation, filtering, dataset intersection
Searching by feature/marker name or by sequence (when BLAST database set up)
Ability to upload custom datasets by drag and dropping Excel templates
User-defined access controls on uploaded datasets, including sharing to groups of users
Dynamic link to Crop Ontology API for QTL trait definition and visualisation
Examples of Pretzel in action
Simultaneously investigating genome to genetic map relationships and zooming to specific regions to visualize genomic features
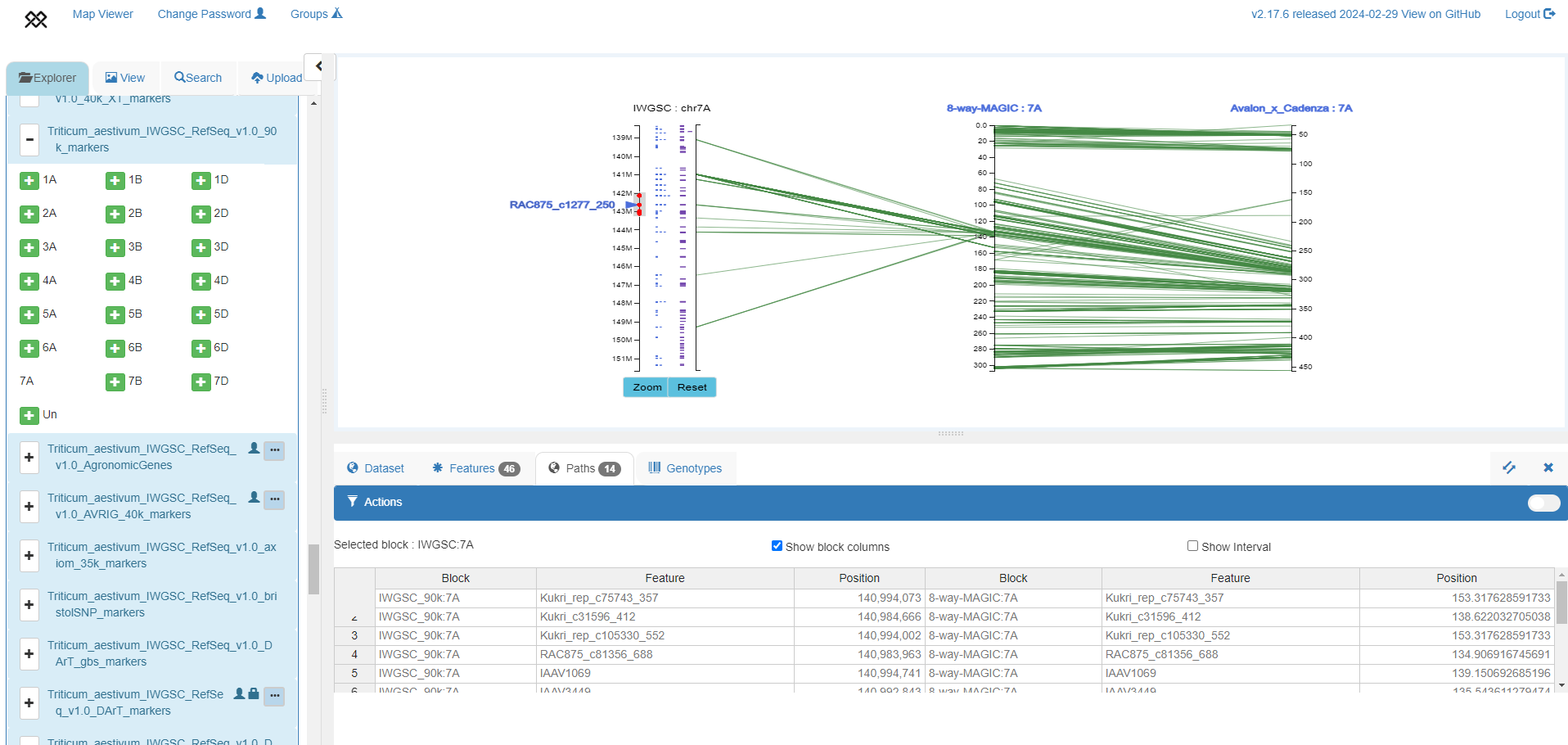
IWGSC RefSeq v1.0 genome assembly chromosome 7A zoomed around the RAC875c1277250 90k marker, with 90k markers (blue) and HC genes (purple) shown (left axis), aligned to the 8-way MAGIC genetic map (Shah et al. 2018, middle axis) and Avalon x Cadenza genetic map (Wang et al. 2014, right axis). The table shows relative position of markers in the genome assembly and MAGIC map.
Visualising chromosome-scale alignments between a genetic map to a genome assembly and syntenic relationships between different species' genomes
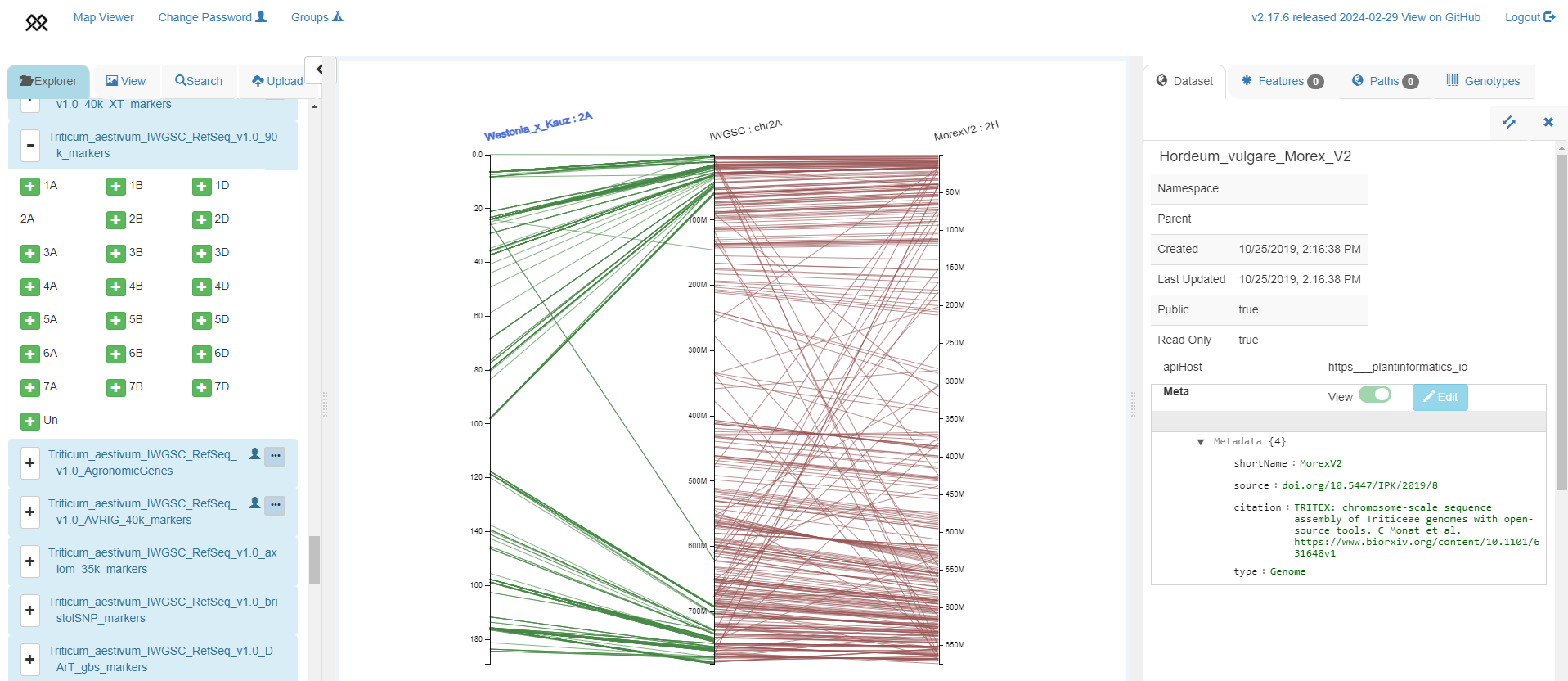
Westonia x Kauz genetic map based on 90k markers (Wang et al. 2014, left axis) aligned to IWGSC RefSeq v1.0 chromosome 2A (middle axis), aligned to barley Morex V2 assembly (right axis) with red lines between orthologous genes between wheat and barley.
Projecting QTL information from a genetic map into a genome assembly
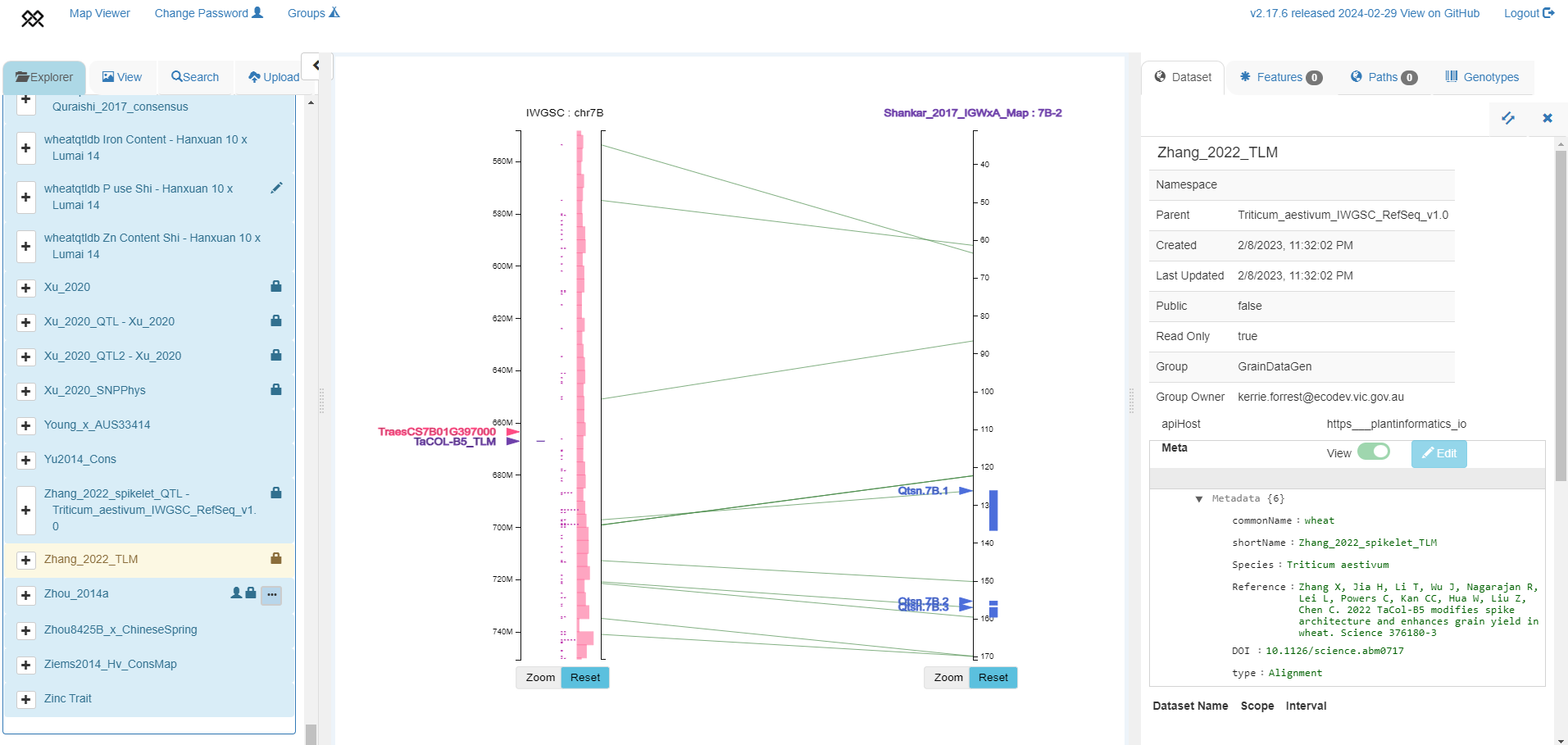
TaCOL-B5 marker position from Zhang et al. 2022 positioned in IWGSC RefSeq v1.0 chromosome 2A (left axis), aligned to SSR map from Shankar et al. 2017 with 3 QTLs shown in blue (right axis).
Exploring haplotypes around a gene in diverse germplasm
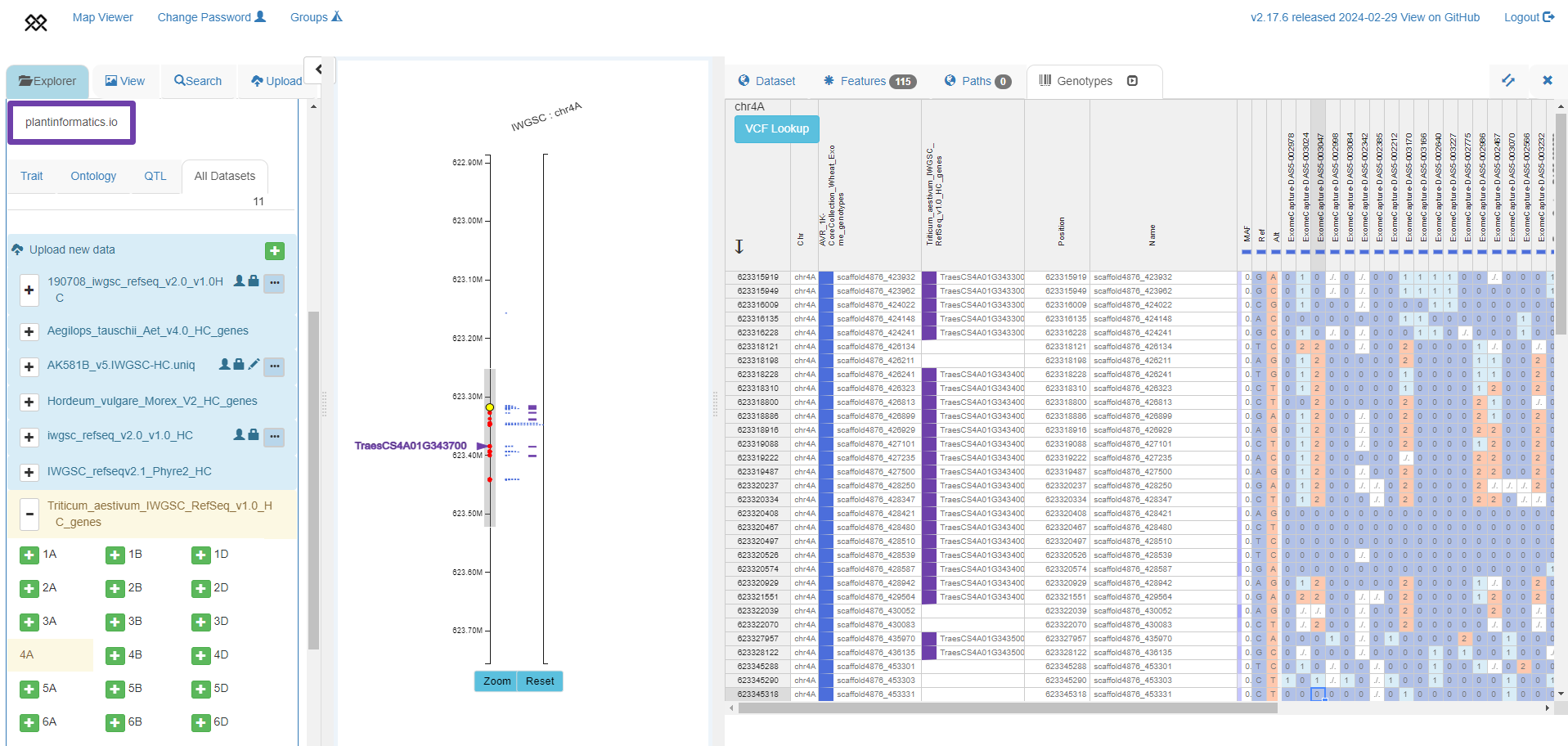
Exome-based haplotypes (right panel) from a subset of accessions from Keeble-Gagnere et al. 2021 (https://doi.org/10.7910/DVN/5LVYI1), with SNP positions shown around TraesCS4A01G343700 in IWGSC RefSeq v1.0 (left panel).
Funding
Currently (2022-) funded as part of the Australian Grains Genebank Strategic Partnership, a $30M joint investment between the Victorian State Government and Grains Research and Development Corporation (GRDC) that aims to unlock the genetic potential of plant genetic resources for the benefit of the Australian grain growers. https://agriculture.vic.gov.au/crops-and-horticulture/the-australian-grains-genebank
Between 2020-2022 funded and developed by Agriculture Victoria, Department of Jobs, Precincts and Regions (DJPR), Victoria, Australia.
Between 2016-2020 funded by the Grains Research Development Corporation (GRDC) and co-developed by Agriculture Victoria and CSIRO, Canberra, Australia.

Owner
- Name: Plant Informatics Collaboration (PICO)
- Login: plantinformatics
- Kind: organization
- Repositories: 7
- Profile: https://github.com/plantinformatics
Collaboration between Agriculture Victoria and CSIRO
Citation (CITATION.cff)
---
cff-version: 1.2.0
message: If you use this software, please cite it using these metadata.
title: Integrating past, present and future wheat research with Pretzel
abstract: Motivation Major advances have been made in the assembly of complex genomes such as wheat. Remaining challenges include linking these new resources to legacy research, as well as lowering the bar to entry for members of the research community who may lack specific bioinformatics skills. Results Pretzel aims to solve these problems by providing an interactive, online environment for data visualisation and analysis which, when loaded with appropriately curated data, can enable researchers with no bioinformatics training to exploit the latest genomic resources. We demonstrate that Pretzel can be used to answer common questions asked by researchers as well as for advanced curation of pseudomolecule structure. Availability Pretzel is implemented in JavaScript and is freely available under a GPLv3 license online at https://github.com/plantinformatics/pretzel. Contact gabriel.keeble-gagnere{at}agriculture.vic.gov.au
authors:
- family-names: Keeble-Gagnère
given-names: Gabriel
orcid: "https://orcid.org/0000-0002-9165-0724"
- family-names: Isdale
given-names: Don
orcid: "https://orcid.org/0000-0002-6553-0075"
- family-names: Suchecki
given-names: Rados-ław
orcid: "https://orcid.org/0000-0003-4992-9497"
- family-names: Kruger
given-names: Alex
- family-names: Lomas
given-names: Kieran
- family-names: Carroll
given-names: David
- family-names: Li
given-names: Sean
- family-names: Whan
given-names: Alex
orcid: "https://orcid.org/0000-0002-6839-2915"
- family-names: Hayden
given-names: Matthew
affiliation: AgriBio Centre for AgriBioscience
- family-names: Tibbits
given-names: Josquin
orcid: "https://orcid.org/0000-0002-4951-7540"
contact:
email : gabriel.keeble-gagnere{at}agriculture.vic.gov.au
version: 2.10.0
date-released: "2021-08-25"
identifiers:
- description: This paper introduces the bioinformatics web application Pretzel.
type: doi
value: "10.1101/517953"
license: GPL-3.0-only
repository-code: "https://github.com/plantinformatics/pretzel"
date-published : "2019/01/01"
keywords :
- genomics
- bioinformatics
- visualisation
- web application
GitHub Events
Total
- Create event: 44
- Release event: 19
- Issues event: 190
- Watch event: 3
- Delete event: 15
- Issue comment event: 184
- Push event: 228
- Pull request review event: 4
- Pull request event: 92
Last Year
- Create event: 44
- Release event: 19
- Issues event: 190
- Watch event: 3
- Delete event: 15
- Issue comment event: 184
- Push event: 228
- Pull request review event: 4
- Pull request event: 92
Committers
Last synced: 8 months ago
Top Committers
| Name | Commits | |
|---|---|---|
| Don | D****e@g****m | 3,295 |
| David Carroll | d****l@c****u | 85 |
| Gabriel Keeble-Gagnere | g****7@g****m | 85 |
| Gabriel Keeble-Gagnere | g****e@e****u | 82 |
| Alex Kruger | a****r@c****u | 56 |
| Kieran Lomas | k****s@c****u | 55 |
| Li, Sean (Data61, Acton) | S****i@c****u | 43 |
| Alex Kruger | A****r@c****u | 36 |
| dependabot[bot] | 4****] | 33 |
| Andrew Spriggs | s****3@y****u | 26 |
| Radoslaw Suchecki | r****i@g****m | 24 |
| kennyAgricultureVic | k****u@a****u | 12 |
| Kendoggs | 3****s | 8 |
| Xi Li | s****2@g****m | 3 |
| Gaire, Raj (Data61, Acton) | R****e@c****u | 3 |
| K L | t****s@g****m | 3 |
| josquint | j****t@u****u | 2 |
| Radoslaw | r****i@a****u | 2 |
| Radoslaw Suchecki | r****i@g****m | 2 |
| David Carroll | d****l@g****m | 2 |
| Kieran Lomas | l****8@s****u | 1 |
Committer Domains (Top 20 + Academic)
Issues and Pull Requests
Last synced: 6 months ago
All Time
- Total issues: 247
- Total pull requests: 111
- Average time to close issues: 3 months
- Average time to close pull requests: 8 months
- Total issue authors: 8
- Total pull request authors: 4
- Average comments per issue: 1.24
- Average comments per pull request: 1.5
- Merged pull requests: 59
- Bot issues: 0
- Bot pull requests: 45
Past Year
- Issues: 102
- Pull requests: 75
- Average time to close issues: 14 days
- Average time to close pull requests: 1 day
- Issue authors: 3
- Pull request authors: 3
- Average comments per issue: 1.12
- Average comments per pull request: 0.07
- Merged pull requests: 50
- Bot issues: 0
- Bot pull requests: 20
Top Authors
Issue Authors
- Don-Isdale (191)
- gabrielkg (28)
- kennyAgricultureVic (11)
- Seanli52 (9)
- rsuchecki (5)
- alexwhan (1)
- Troelsmou (1)
- davidcarroll-csiro (1)
Pull Request Authors
- Don-Isdale (56)
- dependabot[bot] (45)
- kennyAgricultureVic (8)
- Gruek (2)
Top Labels
Issue Labels
Pull Request Labels
Dependencies
- animation-frame ^0.2.5
- antiscroll 8dc9793150
- d3 ^4.10.2
- d3-tip VACLab/d3-tip
- ember-qunit-notifications ^0.1.0
- handsontable ^8
- minimatch isaacs/minimatch#^3.0.4
- pretender ~1.1.0
- 2336 dependencies
- @cablanchard/koelle-sort ^0.1.0 development
- @ember/jquery ^1.1.0 development
- @ember/optional-features ^2.0.0 development
- @faker-js/faker ^6.3.1 development
- @glimmer/component ^1.0.2 development
- @glimmer/tracking ^1.0.2 development
- babel-eslint ^10.1.0 development
- bootstrap-sass ^3.4.1 development
- broccoli-asset-rev ^3.0.0 development
- d3-scale-chromatic ^2.0.0 development
- ember-3x-codemods ^1.12.5 development
- ember-ajax ^5.0.0 development
- ember-auto-import ^1.7.0 development
- ember-bootstrap ^3.1.4 development
- ember-cli 3.22.0 development
- ember-cli-app-version ^4.0.0 development
- ember-cli-babel ^7.23.0 development
- ember-cli-bootstrap-css ^0.2.0 development
- ember-cli-bootstrap-fonts ^0.2.0 development
- ember-cli-dependency-checker ^3.2.0 development
- ember-cli-document-title ^0.4.0 development
- ember-cli-inject-live-reload ^2.0.2 development
- ember-cli-jshint ^2.0.1 development
- ember-cli-release ^1.0.0-beta.2 development
- ember-cli-sass ^10.0.1 development
- ember-cli-sri ^2.1.1 development
- ember-cli-terser ^4.0.1 development
- ember-cli-test-loader ^3.0.0 development
- ember-component-inbound-actions ^1.3.1 development
- ember-content-editable 1.0.5 development
- ember-data ^3.22.1 development
- ember-data-model-fragments ^4.0.0 development
- ember-export-application-global ^2.0.1 development
- ember-fetch ^8.0.2 development
- ember-file-upload ^3.0.5 development
- ember-load-initializers ^2.1.2 development
- ember-local-storage ^1.7.2 development
- ember-maybe-import-regenerator ^0.1.6 development
- ember-modal-dialog ^3.0.3 development
- ember-qunit ^4.6.0 development
- ember-raf-scheduler ^0.1.0 development
- ember-resolver ^8.0.2 development
- ember-scroll-to 0.6.5 development
- ember-simple-auth ^3.0.1 development
- ember-spectrum-color-picker ^0.10.1 development
- ember-split-view-modifier github:plantinformatics/ember-split-view-modifier development
- ember-template-lint ^2.14.0 development
- ember-tether ^2.0.1 development
- ember-welcome-page ^4.0.0 development
- eslint ^7.13.0 development
- eslint-plugin-ember ^9.6.0 development
- eslint-plugin-node ^11.1.0 development
- jsoneditor ^9.1.2 development
- loader.js ^4.7.0 development
- npm-run-all ^4.1.5 development
- qunit-dom ^1.6.0 development
- sass ^1.29.0 development
- caniuse-lite ^1.0.30001230
- colresizable github:felipedgarcia/colresizable
- ember-array-helper >=5
- ember-cli-build-date github:bracke/ember-cli-build-date
- ember-cli-htmlbars ^5.3.1
- ember-cli-multi-store-service git+https://git@github.com/plantinformatics/ember-cli-multi-store-service.git
- ember-composable-helpers ^4.3.1
- ember-concurrency ^1.3.0
- ember-contextual-table ^4
- ember-csv ^1.0.2
- ember-jsoneditor git+https://git@github.com/plantinformatics/ember-jsoneditor.git
- ember-lodash ^4.19.5
- ember-promise-helpers ^1.0.9
- ember-radio-button ^2.0.1
- ember-raf-scheduler
- ember-sortable
- ember-source ^3.22.1
- ember-toggle ^7.0.0
- ember-tooltips ^3.4.5
- ember-truth-helpers ^3.0.0
- ember-wormhole ^0.6.0
- interval-tree-1d github:mikolalysenko/interval-tree-1d
- moment ^2.29.1
- node-sass ^5.0.0
- normalize-wheel ^1.0.1
- numeric github:lukaiser/numeric
- phantom ^6.3.0
- 843 dependencies
- @loopback/build ^8.1.0 development
- @loopback/eslint-config ^12.0.2 development
- @loopback/testlab ^4.1.0 development
- @types/node ^12.20.43 development
- chai ^4.3.6 development
- eslint ^8.9.0 development
- eslint-config-loopback ^13.1.0 development
- eventsource ^1.1.0 development
- mocha ^9.2.1 development
- nodemon ^2.0.15 development
- nsp ^2.1.0 development
- qs ^6.10.3 development
- smtp-server ^3.9.0 development
- source-map-support ^0.5.21 development
- superagent ^7.1.1 development
- typescript ~4.5.5 development
- @loopback/boot ^4.1.0
- @loopback/booter-lb3app ^3.1.0
- @loopback/core ^3.1.0
- @loopback/repository ^4.1.0
- @loopback/rest ^11.1.0
- @loopback/rest-explorer ^4.1.0
- @loopback/service-proxy ^4.1.0
- async ^3.2.3
- bent ^7.3.12
- body-parser ^1.19.2
- compression ^1.7.4
- cors ^2.8.5
- dotenv ^16.0.0
- express-sse ^0.5.3
- helmet ^5.0.2
- jquery-param ^1.1.8
- lodash ^4.17.21
- loopback ^3.28.0
- loopback-boot ^3.3.1
- loopback-component-explorer ^6.5.1
- loopback-component-passport ^3.12.0
- loopback-connector-mongodb ^6.1.0
- loopback-datasource-juggler ^4.27.0
- memory-cache ^0.2.0
- mongoose ^6.2.3
- morgan ^1.10.0
- node ^17.4.0
- passport-ldapauth 3.0.1
- promise-queue ^2.2.5
- serve-favicon ^2.5.0
- tslib ^2.0.0


