Science Score: 57.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
✓CITATION.cff file
Found CITATION.cff file -
✓codemeta.json file
Found codemeta.json file -
✓.zenodo.json file
Found .zenodo.json file -
✓DOI references
Found 13 DOI reference(s) in README -
○Academic publication links
-
○Academic email domains
-
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (10.7%) to scientific vocabulary
Keywords
Repository
Nextflow version of Sopa - spatial omics pipeline and analysis
Basic Info
- Host: GitHub
- Owner: nf-core
- License: mit
- Language: Nextflow
- Default Branch: dev
- Homepage: https://nf-co.re/sopa/dev/
- Size: 478 KB
Statistics
- Stars: 7
- Watchers: 3
- Forks: 1
- Open Issues: 1
- Releases: 0
Topics
Metadata Files
README.md

Introduction
nf-core/sopa is the Nextflow version of Sopa. Built on top of SpatialData, Sopa enables processing and analyses of spatial omics data with single-cell resolution (spatial transcriptomics or multiplex imaging data) using a standard data structure and output. We currently support the following technologies: Xenium, Visium HD, MERSCOPE, CosMX, PhenoCycler, MACSima, Molecural Cartography, and others. It outputs a .zarr directory containing a processed SpatialData object, and a .explorer directory for visualization.
[!WARNING] If you are interested in the main Sopa python package, refer to this Sopa repository. Else, if you want to use Nextflow, you are in the good place.
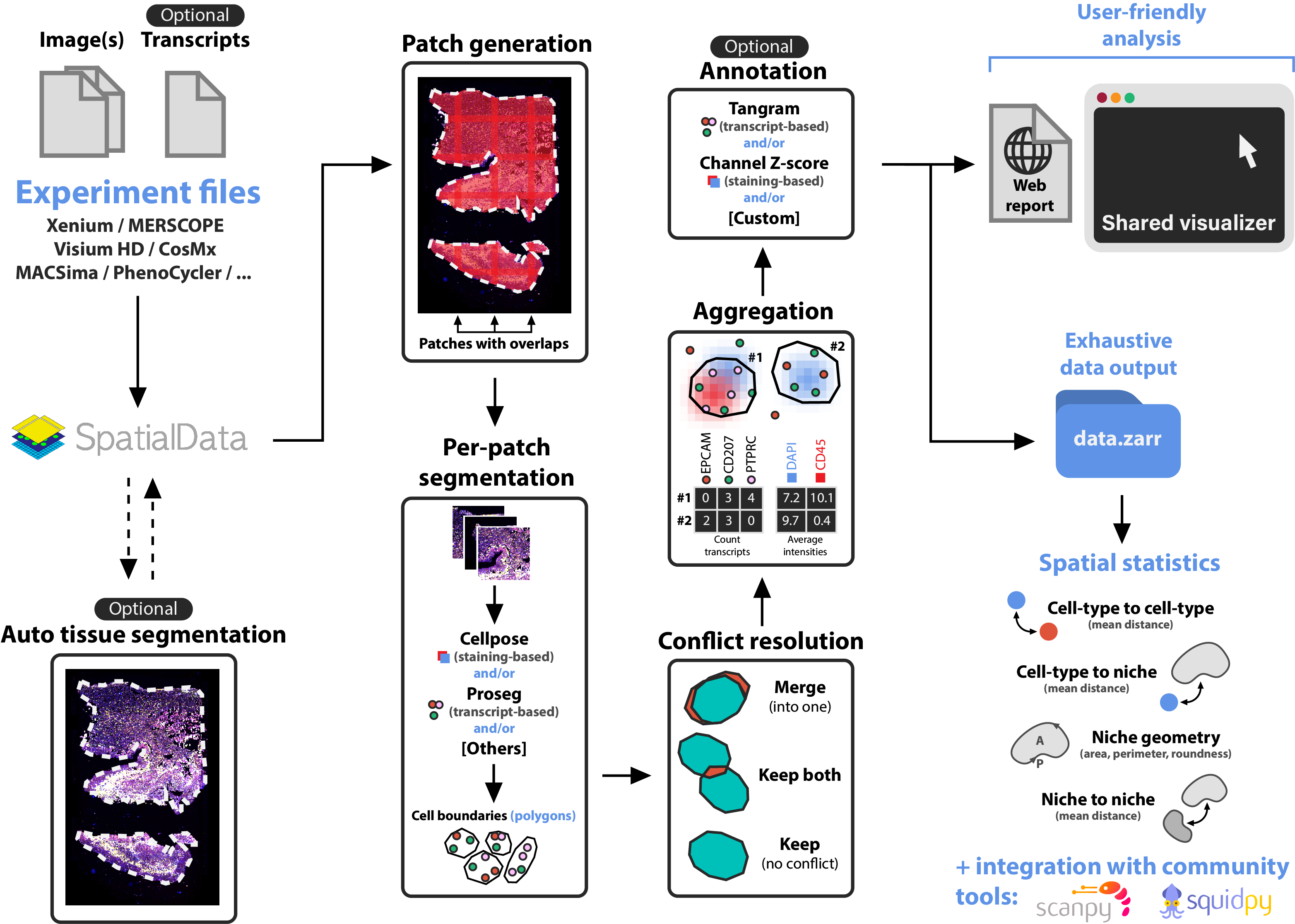
- (Visium HD only) Raw data processing with Space Ranger
- (Optional) Tissue segmentation
- Cell segmentation with Cellpose, Baysor, Proseg, Comseg, Stardist, ...
- Aggregation, i.e. counting the transcripts inside the cells and/or averaging the channel intensities inside cells
- (Optional) Cell-type annotation
- User-friendly output creation for visualization and quick analysis
- Full SpatialData object export as a
.zarrdirectory
After running nf-core/sopa, you can continue analyzing your SpatialData object with sopa as a Python package.
Usage
[!NOTE] If you are new to Nextflow and nf-core, please refer to this page on how to set-up Nextflow. Make sure to test your setup with
-profile testbefore running the workflow on actual data.
First, prepare a samplesheet that lists the data_path to each sample data directory (typically, the per-sample output of the Xenium/MERSCOPE/etc, see more info here). You can optionally add sample to provide a name to your output directory, else it will be named based on data_path. Here is a samplesheet example:
samplesheet.csv:
csv
sample,data_path
SAMPLE1,/path/to/one/merscope_directory
SAMPLE2,/path/to/one/merscope_directory
[!WARNING] If you have Visium HD data, the samplesheet will have a different format than the one above. Directly refer to the usage documentation and the parameter documentation.
Then, choose the Sopa parameters. You can find existing Sopa params files here, and follow the corresponding README instructions of to get your -params-file argument.
Now, you can run the pipeline using:
bash
nextflow run nf-core/sopa \
-profile <docker/singularity/.../institute> \
--input samplesheet.csv \
-params-file <PARAMS_FILE> \
--outdir <OUTDIR>
[!WARNING] Please provide pipeline parameters via the CLI or Nextflow
-params-fileoption. Custom config files including those provided by the-cNextflow option can be used to provide any configuration except for parameters; see docs.
For more details and further functionality, please refer to the usage documentation and the parameter documentation.
Pipeline output
To see the results of an example test run with a full size dataset refer to the results tab on the nf-core website pipeline page. For more details about the output files and reports, please refer to the output documentation.
Credits
nf-core/sopa was originally written by Quentin Blampey during his work at the following institutions: CentraleSupélec, Gustave Roussy Institute, and Université Paris-Saclay.
We thank the following people for their extensive assistance in the development of this pipeline:
Contributions and Support
If you would like to contribute to this pipeline, please see the contributing guidelines.
For further information or help, don't hesitate to get in touch on the Slack #sopa channel (you can join with this invite).
Citations
An extensive list of references for the tools used by the pipeline can be found in the CITATIONS.md file.
You can cite the sopa publication as follows:
Sopa: a technology-invariant pipeline for analyses of image-based spatial omics.
Quentin Blampey, Kevin Mulder, Margaux Gardet, Stergios Christodoulidis, Charles-Antoine Dutertre, Fabrice André, Florent Ginhoux & Paul-Henry Cournède.
Nat Commun. 2024 June 11. doi: 10.1038/s41467-024-48981-z
You can cite the nf-core publication as follows:
The nf-core framework for community-curated bioinformatics pipelines.
Philip Ewels, Alexander Peltzer, Sven Fillinger, Harshil Patel, Johannes Alneberg, Andreas Wilm, Maxime Ulysse Garcia, Paolo Di Tommaso & Sven Nahnsen.
Nat Biotechnol. 2020 Feb 13. doi: 10.1038/s41587-020-0439-x.
Owner
- Name: nf-core
- Login: nf-core
- Kind: organization
- Email: core@nf-co.re
- Website: http://nf-co.re
- Twitter: nf_core
- Repositories: 84
- Profile: https://github.com/nf-core
A community effort to collect a curated set of analysis pipelines built using Nextflow.
Citation (CITATIONS.md)
# nf-core/sopa: Citations ## [sopa](https://www.nature.com/articles/s41467-024-48981-z) > Blampey, Q., Mulder, K., Gardet, M. et al. Sopa: a technology-invariant pipeline for analyses of image-based spatial omics. Nat Commun 15, 4981 (2024). https://doi.org/10.1038/s41467-024-48981-z ## [nf-core](https://pubmed.ncbi.nlm.nih.gov/32055031/) > Ewels PA, Peltzer A, Fillinger S, Patel H, Alneberg J, Wilm A, Garcia MU, Di Tommaso P, Nahnsen S. The nf-core framework for community-curated bioinformatics pipelines. Nat Biotechnol. 2020 Mar;38(3):276-278. doi: 10.1038/s41587-020-0439-x. PubMed PMID: 32055031. ## [Nextflow](https://pubmed.ncbi.nlm.nih.gov/28398311/) > Di Tommaso P, Chatzou M, Floden EW, Barja PP, Palumbo E, Notredame C. Nextflow enables reproducible computational workflows. Nat Biotechnol. 2017 Apr 11;35(4):316-319. doi: 10.1038/nbt.3820. PubMed PMID: 28398311. ## Pipeline tools - [AnnData](https://github.com/scverse/anndata) > Virshup I, Rybakov S, Theis FJ, Angerer P, Wolf FA. bioRxiv 2021.12.16.473007; doi: https://doi.org/10.1101/2021.12.16.473007 - [Scanpy](https://github.com/theislab/scanpy) > Wolf F, Angerer P, Theis F. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 19, 15 (2018). doi: https://doi.org/10.1186/s13059-017-1382-0 - [Space Ranger](https://www.10xgenomics.com/support/software/space-ranger) > 10x Genomics Space Ranger 2.1.0 [Online] - [SpatialData](https://www.biorxiv.org/content/10.1101/2023.05.05.539647v1) > Marconato L, Palla G, Yamauchi K, Virshup I, Heidari E, Treis T, Toth M, Shrestha R, Vöhringer H, Huber W, Gerstung M, Moore J, Theis F, Stegle O. SpatialData: an open and universal data framework for spatial omics. bioRxiv 2023.05.05.539647; doi: https://doi.org/10.1101/2023.05.05.539647 ## Software packaging/containerisation tools - [Anaconda](https://anaconda.com) > Anaconda Software Distribution. Computer software. Vers. 2-2.4.0. Anaconda, Nov. 2016. Web. - [Bioconda](https://pubmed.ncbi.nlm.nih.gov/29967506/) > Grüning B, Dale R, Sjödin A, Chapman BA, Rowe J, Tomkins-Tinch CH, Valieris R, Köster J; Bioconda Team. Bioconda: sustainable and comprehensive software distribution for the life sciences. Nat Methods. 2018 Jul;15(7):475-476. doi: 10.1038/s41592-018-0046-7. PubMed PMID: 29967506. - [BioContainers](https://pubmed.ncbi.nlm.nih.gov/28379341/) > da Veiga Leprevost F, Grüning B, Aflitos SA, Röst HL, Uszkoreit J, Barsnes H, Vaudel M, Moreno P, Gatto L, Weber J, Bai M, Jimenez RC, Sachsenberg T, Pfeuffer J, Alvarez RV, Griss J, Nesvizhskii AI, Perez-Riverol Y. BioContainers: an open-source and community-driven framework for software standardization. Bioinformatics. 2017 Aug 15;33(16):2580-2582. doi: 10.1093/bioinformatics/btx192. PubMed PMID: 28379341; PubMed Central PMCID: PMC5870671. - [Docker](https://dl.acm.org/doi/10.5555/2600239.2600241) > Merkel, D. (2014). Docker: lightweight linux containers for consistent development and deployment. Linux Journal, 2014(239), 2. doi: 10.5555/2600239.2600241. - [Singularity](https://pubmed.ncbi.nlm.nih.gov/28494014/) > Kurtzer GM, Sochat V, Bauer MW. Singularity: Scientific containers for mobility of compute. PLoS One. 2017 May 11;12(5):e0177459. doi: 10.1371/journal.pone.0177459. eCollection 2017. PubMed PMID: 28494014; PubMed Central PMCID: PMC5426675.
GitHub Events
Total
- Watch event: 4
- Delete event: 3
- Issue comment event: 8
- Push event: 36
- Pull request review event: 4
- Pull request review comment event: 3
- Pull request event: 8
- Fork event: 1
- Create event: 5
Last Year
- Watch event: 4
- Delete event: 3
- Issue comment event: 8
- Push event: 36
- Pull request review event: 4
- Pull request review comment event: 3
- Pull request event: 8
- Fork event: 1
- Create event: 5
Issues and Pull Requests
Last synced: 7 months ago
All Time
- Total issues: 0
- Total pull requests: 1
- Average time to close issues: N/A
- Average time to close pull requests: N/A
- Total issue authors: 0
- Total pull request authors: 1
- Average comments per issue: 0
- Average comments per pull request: 0.0
- Merged pull requests: 0
- Bot issues: 0
- Bot pull requests: 0
Past Year
- Issues: 0
- Pull requests: 1
- Average time to close issues: N/A
- Average time to close pull requests: N/A
- Issue authors: 0
- Pull request authors: 1
- Average comments per issue: 0
- Average comments per pull request: 0.0
- Merged pull requests: 0
- Bot issues: 0
- Bot pull requests: 0
Top Authors
Issue Authors
Pull Request Authors
- quentinblampey (5)
Top Labels
Issue Labels
Pull Request Labels
Dependencies
- actions/upload-artifact v4 composite
- octokit/request-action v2.x composite
- seqeralabs/action-tower-launch v2 composite
- actions/upload-artifact v4 composite
- seqeralabs/action-tower-launch v2 composite
- mshick/add-pr-comment b8f338c590a895d50bcbfa6c5859251edc8952fc composite
- actions/checkout 11bd71901bbe5b1630ceea73d27597364c9af683 composite
- conda-incubator/setup-miniconda a4260408e20b96e80095f42ff7f1a15b27dd94ca composite
- eWaterCycle/setup-apptainer main composite
- jlumbroso/free-disk-space 54081f138730dfa15788a46383842cd2f914a1be composite
- nf-core/setup-nextflow v2 composite
- actions/stale 28ca1036281a5e5922ead5184a1bbf96e5fc984e composite
- actions/setup-python 0b93645e9fea7318ecaed2b359559ac225c90a2b composite
- eWaterCycle/setup-apptainer 4bb22c52d4f63406c49e94c804632975787312b3 composite
- jlumbroso/free-disk-space 54081f138730dfa15788a46383842cd2f914a1be composite
- nf-core/setup-nextflow v2 composite
- actions/checkout 11bd71901bbe5b1630ceea73d27597364c9af683 composite
- actions/setup-python 0b93645e9fea7318ecaed2b359559ac225c90a2b composite
- peter-evans/create-or-update-comment 71345be0265236311c031f5c7866368bd1eff043 composite
- actions/checkout 11bd71901bbe5b1630ceea73d27597364c9af683 composite
- actions/setup-python 0b93645e9fea7318ecaed2b359559ac225c90a2b composite
- actions/upload-artifact b4b15b8c7c6ac21ea08fcf65892d2ee8f75cf882 composite
- nf-core/setup-nextflow v2 composite
- pietrobolcato/action-read-yaml 1.1.0 composite
- dawidd6/action-download-artifact 80620a5d27ce0ae443b965134db88467fc607b43 composite
- marocchino/sticky-pull-request-comment 331f8f5b4215f0445d3c07b4967662a32a2d3e31 composite
- actions/setup-python 0b93645e9fea7318ecaed2b359559ac225c90a2b composite
- rzr/fediverse-action master composite
- zentered/bluesky-post-action 80dbe0a7697de18c15ad22f4619919ceb5ccf597 composite
- actions/checkout 11bd71901bbe5b1630ceea73d27597364c9af683 composite
- mshick/add-pr-comment b8f338c590a895d50bcbfa6c5859251edc8952fc composite
- nichmor/minimal-read-yaml v0.0.2 composite












