gosling
A declarative interactive genomics visualization library for Python.
Science Score: 54.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
✓CITATION.cff file
Found CITATION.cff file -
✓codemeta.json file
Found codemeta.json file -
✓.zenodo.json file
Found .zenodo.json file -
○DOI references
-
○Academic publication links
-
✓Committers with academic emails
2 of 5 committers (40.0%) from academic institutions -
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (15.2%) to scientific vocabulary
Keywords
Repository
A declarative interactive genomics visualization library for Python.
Basic Info
- Host: GitHub
- Owner: gosling-lang
- License: mit
- Language: Python
- Default Branch: main
- Homepage: https://gosling-lang.github.io/gos
- Size: 13 MB
Statistics
- Stars: 229
- Watchers: 3
- Forks: 16
- Open Issues: 13
- Releases: 9
Topics
Metadata Files
README.md
gos 🦆
gos is a declarative genomics visualization library for Python. It is built on top of the Gosling JSON specification, providing a simplified interface for authoring interactive genomic visualizations.
Installation
The gos API is under active development. Feedback is appreciated and welcomed.
bash
pip install gosling[all]
Documentation
See the Documentation Site for more information.
Example
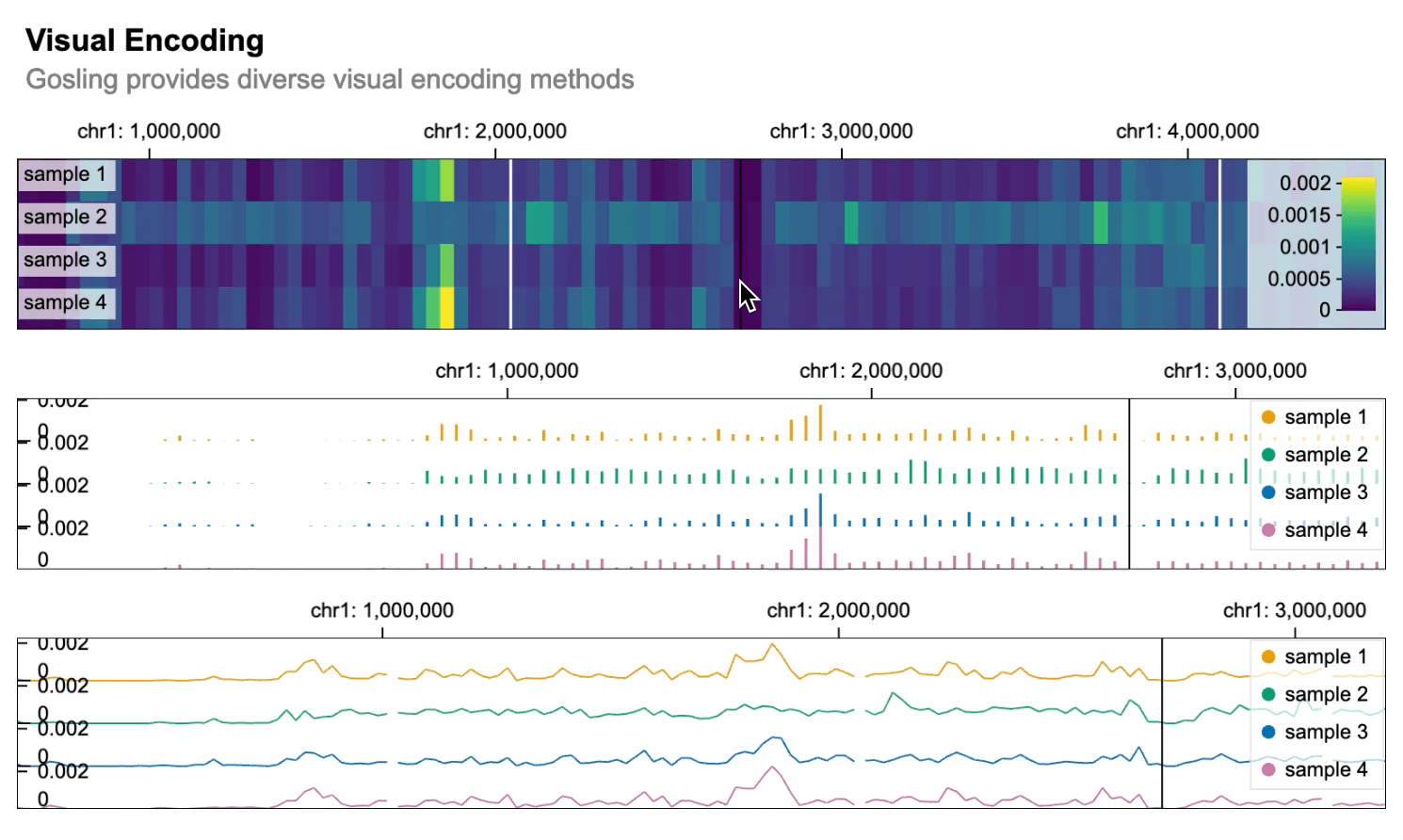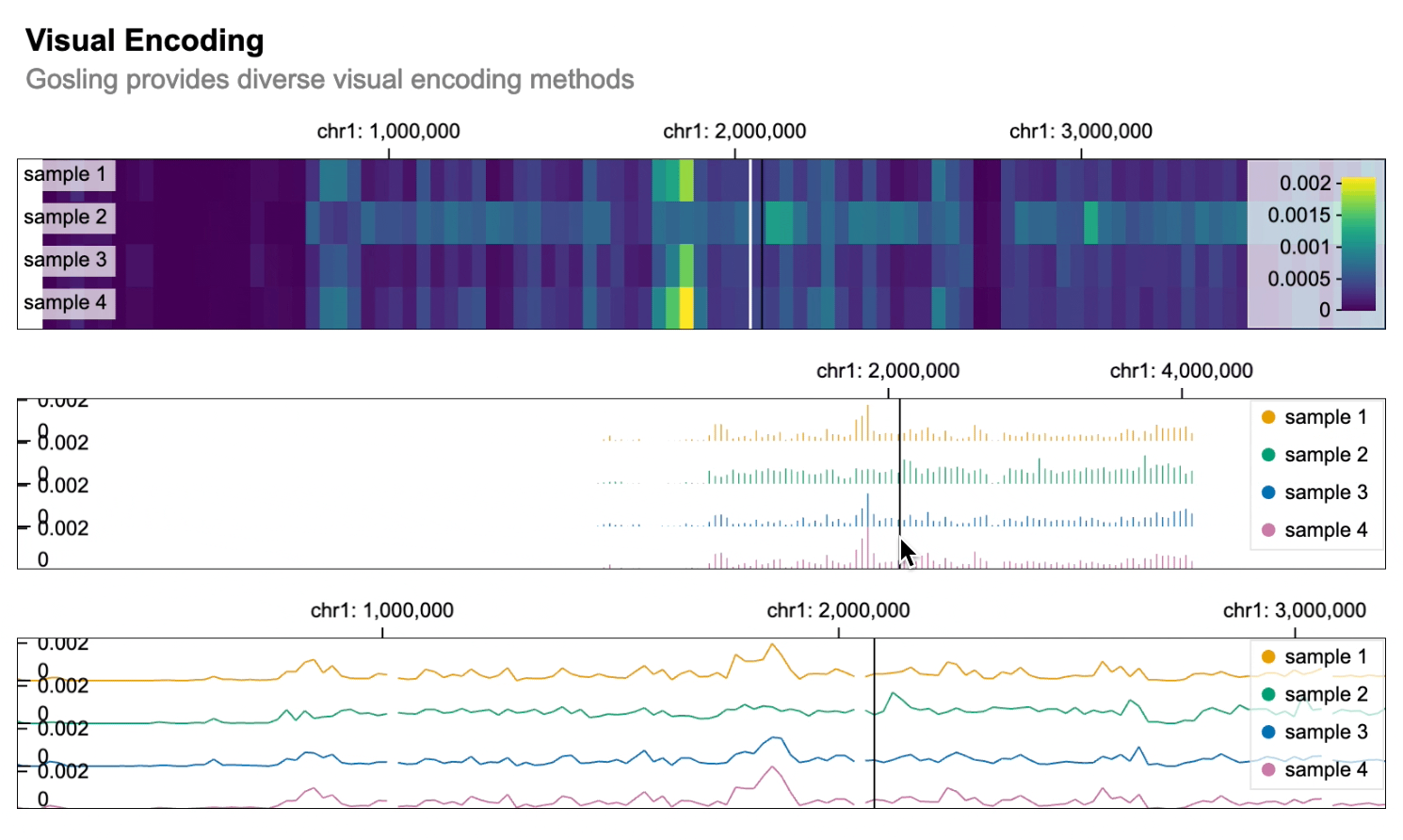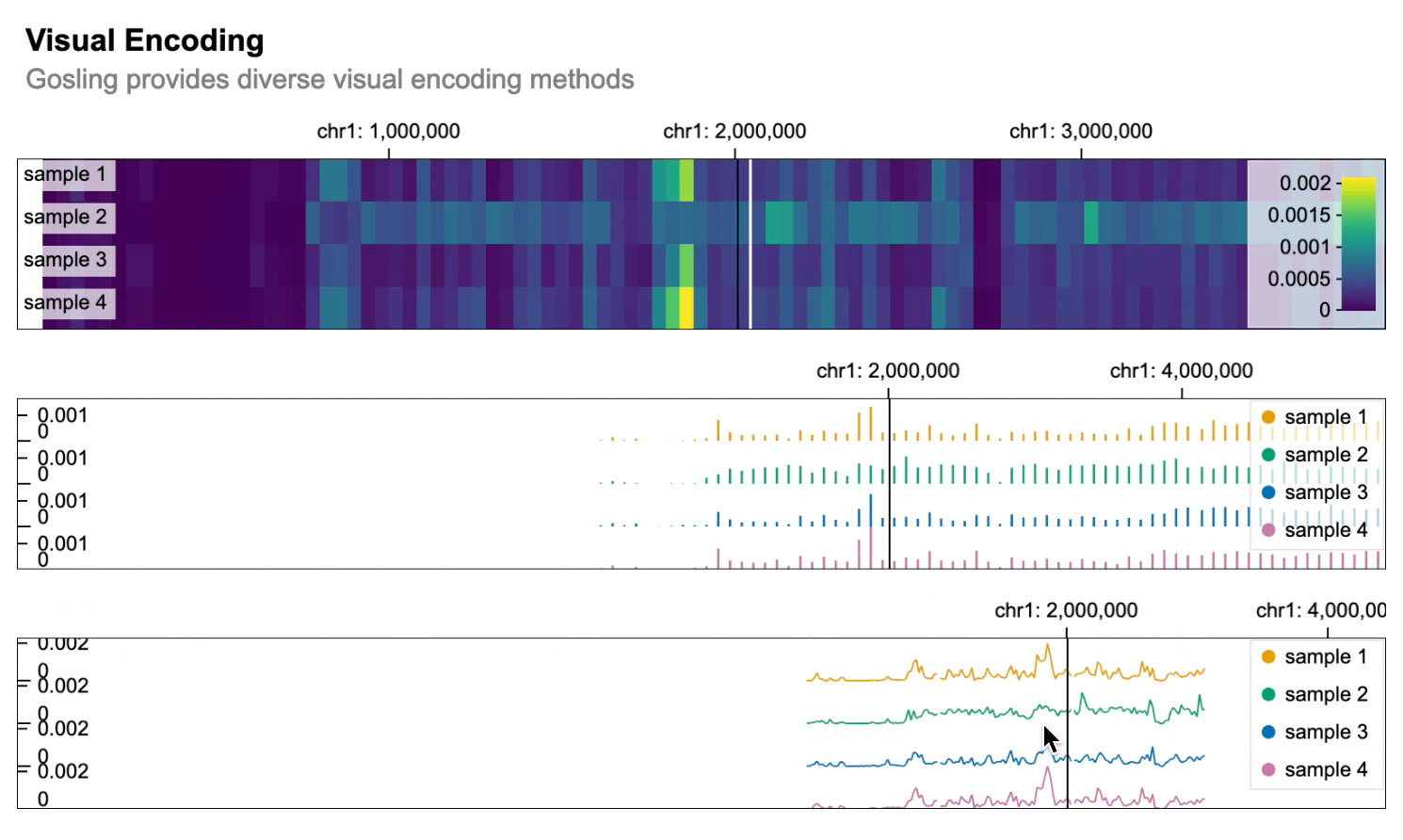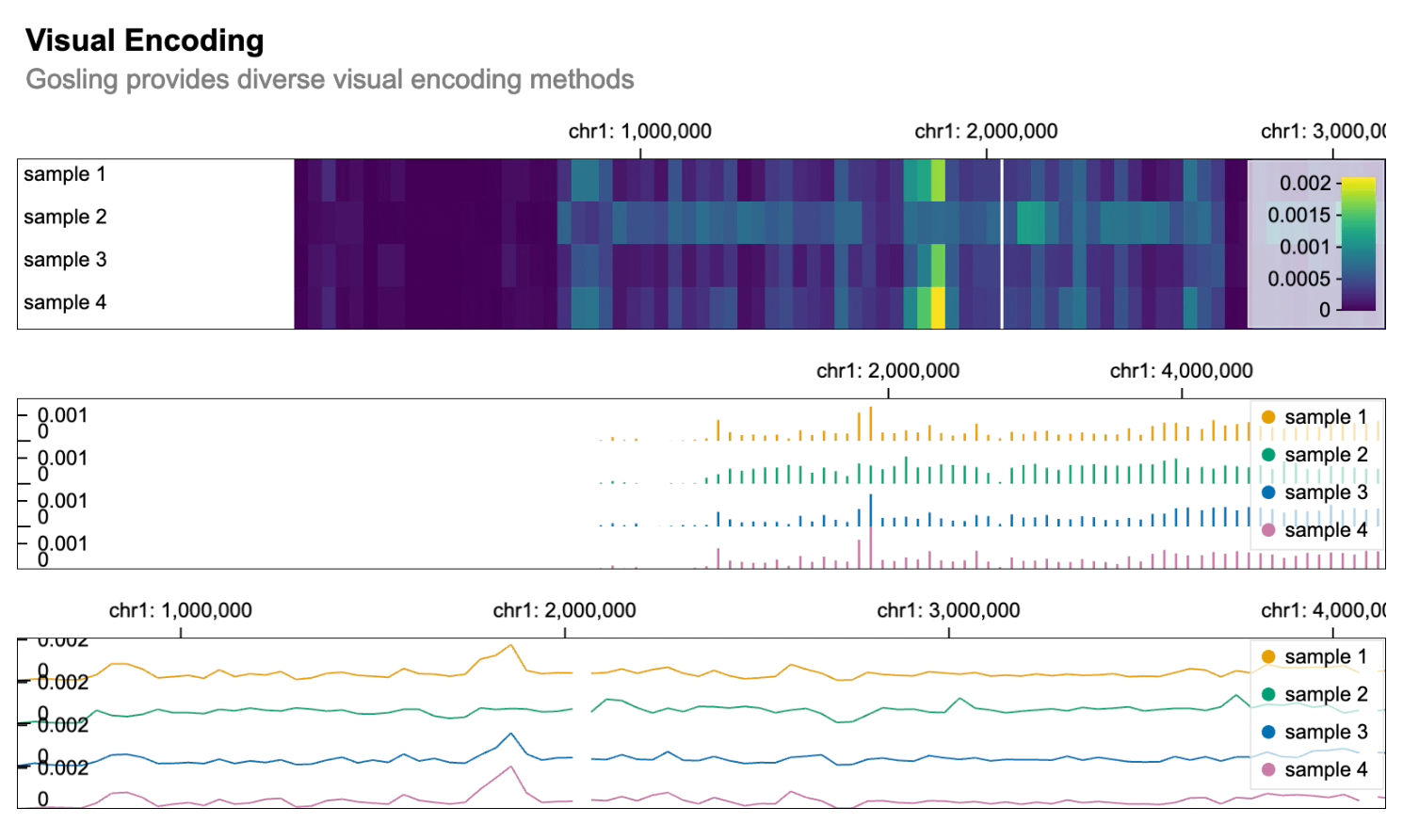
```python import gosling as gos
data = gos.multivec( url="https://server.gosling-lang.org/api/v1/tileset_info/?d=cistrome-multivec", row="sample", column="position", value="peak", categories=["sample 1", "sample 2", "sample 3", "sample 4"], binSize=5, )
base_track = gos.Track(data, width=800, height=100)
heatmap = basetrack.markrect().encode( x=gos.X("start:G", axis="top"), xe="end:G", row=gos.Row("sample:N", legend=True), color=gos.Color("peak:Q", legend=True), )
bars = basetrack.markbar().encode( x=gos.X("position:G", axis="top"), y="peak:Q", row="sample:N", color=gos.Color("sample:N", legend=True), )
lines = basetrack.markline().encode( x=gos.X("position:G", axis="top"), y="peak:Q", row="sample:N", color=gos.Color("sample:N", legend=True), )
gos.vertical(heatmap, bars, lines).properties( title="Visual Encoding", subtitle="Gosling provides diverse visual encoding methods", layout="linear", centerRadius=0.8, xDomain=gos.GenomicDomain(chromosome="1", interval=[1, 3000500]), ) ```
Example Gallery
We have started a
gallery of community
examples in gosling/examples/. If you are interested in contributing, please
feel free to submit a PR! Checkout the
existing JSON examples if you are looking
for inspiration.
Changelog
Check the GitHub Releases for a detailed changelog.
Development
The source code for gos is a hybrid of Python and TypeScript (used for the anywidget component). It requires both:
Please ensure both are installed before proceeding.
Tests
Run the test suite with:
sh
uv run pytest
Notebooks
To try out the library in the example notebooks/, launch Jupyter Lab with:
sh
uv run jupyter lab
Widget
The widgets implementation is split between ./gosling/_widget.py (the Python
component) and ./frontend/widget.ts (the TypeScript component).
To modify the widget's behavior in the front end, edit ./frontend/widget.ts
and compile with:
sh
deno task build
Use deno task dev to watch for changes and recompile automatically.
Docs
To build and preview the documentation locally:
sh
uv run docs/build.py
uv run python -m http.server -d docs/dist
Open your browser to http://localhost:8000 to view the generated docs.
Auto-generate Schema Bindings
Much of the Python code in this repository is automatically generated from the
Gosling schema to keep the bindings in sync. This includes both the bindings in
gosling/schema/ and the corresponding API documentation in
doc/user_guide/API.rst.
Do not edit these files manually. Instead, regenerate them using:
```sh
Update gosling/schema/*
uv run tools/generateschemawrapper.py
Update API docs
uv run tools/generateapidocs.py ```
Use a tag_name that corresponds to a valid
Gosling.js Release (e.g.,
v0.12.3).
You must commit the changes and create a new release. Schema updates usually require at least a minor version bump, but the exact versioning is up to the maintainer.
Release
Releases are managed via the GitHub UI. The release tag determines the package version published to PyPI.
- Create a tag
- Click "Choose a tag", then type a new tag in the format
v[major].[minor].[patch]to create it. - Note: The UI is not obvious about this. You can create a tag here, not just select one.
- Generate release notes
- Click "Generate Release Notes" to auto-summarize changes from merged PRs.
- Edit to exclude irrelevant changes for end users (e.g., docs or CI).
- Document significant changes
- Add migration steps or noteworthy updates.
- Ensure PR titles are clear and consistent.
- Publish the release
- Click Publish release to make it public.
- This triggers a workflow that builds the package and publishes it to PyPI using the new tag.
Design & Implementation
gos is inspired by and borrows heavily from Altair both in project philosophy
and implementation. The internal Python API is auto-generated from the Gosling
specification using code adapted directly from Altair to generate Vega-Lite
bindings. This design choice guarantees that visualizations are type-checked in
complete concordance with the Gosling specification, and that the Python API
remains consistent with the evolving schema over time. Special thanks to
Jake Vanderplas and others on
schemapi.
Owner
- Name: Gosling
- Login: gosling-lang
- Kind: organization
- Location: United States of America
- Website: https://gosling.js.org
- Repositories: 23
- Profile: https://github.com/gosling-lang
The data visualization grammar of scalable linked interactive nucleotide graphics. A project of the Gehlenborg Lab at @hms-dbmi.
Citation (CITATION.cff)
cff-version: 1.2.0
message: "If you use this software, please cite it using these metadata."
title: "Gos"
authors:
- given-names: "Trevor"
family-names: "Manz"
orcid: "https://orcid.org/0000-0001-7694-5164"
affiliation: "Harvard Medical School"
- given-names: "Sehi"
family-names: "L'Yi"
orcid: "https://orcid.org/0000-0001-7720-2848"
affiliation: "Harvard Medical School"
url: "https://github.com/gosling-lang/gos"
preferred-citation:
type: article
title: "Gos: a declarative library for interactive genomics visualization in Python"
authors:
- given-names: "Trevor"
family-names: "Manz"
orcid: "https://orcid.org/0000-0001-7694-5164"
affiliation: "Harvard Medical School"
orcid: "https://orcid.org/0000-0001-9198-5498"
- given-names: "Sehi"
family-names: "L'Yi"
orcid: "https://orcid.org/0000-0001-7720-2848"
affiliation: "Harvard Medical School"
- given-names: "Nils"
family-names: "Gehlenborg"
affiliation: "Harvard Medical School"
orcid: "https://orcid.org/0000-0003-0327-8297"
doi: "10.31219/osf.io/yn3ce"
journal: "OSF Preprints"
month: 6
year: 2022
GitHub Events
Total
- Create event: 20
- Release event: 4
- Issues event: 4
- Watch event: 18
- Delete event: 15
- Issue comment event: 12
- Push event: 34
- Pull request review event: 3
- Pull request review comment event: 2
- Pull request event: 29
- Fork event: 2
Last Year
- Create event: 20
- Release event: 4
- Issues event: 4
- Watch event: 18
- Delete event: 15
- Issue comment event: 12
- Push event: 34
- Pull request review event: 3
- Pull request review comment event: 2
- Pull request event: 29
- Fork event: 2
Committers
Last synced: almost 3 years ago
All Time
- Total Commits: 146
- Total Committers: 5
- Avg Commits per committer: 29.2
- Development Distribution Score (DDS): 0.144
Top Committers
| Name | Commits | |
|---|---|---|
| Trevor Manz | t****z@g****m | 125 |
| Sehi L'Yi | s****i@g****m | 15 |
| Sehi L'Yi | s****i@h****u | 4 |
| Nils Gehlenborg | n****s@h****u | 1 |
| James Scott-Brown | j****s@j****m | 1 |
Committer Domains (Top 20 + Academic)
Issues and Pull Requests
Last synced: 6 months ago
All Time
- Total issues: 1
- Total pull requests: 13
- Average time to close issues: about 17 hours
- Average time to close pull requests: 8 days
- Total issue authors: 1
- Total pull request authors: 3
- Average comments per issue: 2.0
- Average comments per pull request: 0.54
- Merged pull requests: 7
- Bot issues: 0
- Bot pull requests: 7
Past Year
- Issues: 1
- Pull requests: 13
- Average time to close issues: about 17 hours
- Average time to close pull requests: 8 days
- Issue authors: 1
- Pull request authors: 3
- Average comments per issue: 2.0
- Average comments per pull request: 0.54
- Merged pull requests: 7
- Bot issues: 0
- Bot pull requests: 7
Top Authors
Issue Authors
- leannmlindsey (1)
- alexvpickering (1)
- tyronechen (1)
- violacimatti (1)
- EnrichettaMileti (1)
Pull Request Authors
- manzt (13)
- dependabot[bot] (5)
- gosling-bot[bot] (2)
- colobas (1)
Top Labels
Issue Labels
Pull Request Labels
Packages
- Total packages: 1
-
Total downloads:
- pypi 282 last-month
- Total dependent packages: 0
- Total dependent repositories: 2
- Total versions: 22
- Total maintainers: 1
pypi.org: gosling
Python bindings to generate Gosling visualizations
- Documentation: https://gosling.readthedocs.io/
- License: MIT
-
Latest release: 0.3.0
published 10 months ago
Rankings
Maintainers (1)
Dependencies
- actions/checkout v4 composite
- actions/setup-python v4 composite
- actions/checkout v4 composite
- actions/deploy-pages main composite
- actions/setup-python v4 composite
- actions/upload-pages-artifact v2 composite
- actions/checkout v4 composite
- actions/setup-python v4 composite
- pypa/gh-action-pypi-publish release/v1 composite
- jinja2 *
- jsonschema >=3.0
- pandas >=1.0,<2.0

