cutevariant
A standalone and free application to explore genetics variations from VCF file
Science Score: 54.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
✓CITATION.cff file
Found CITATION.cff file -
✓codemeta.json file
Found codemeta.json file -
✓.zenodo.json file
Found .zenodo.json file -
○DOI references
-
○Academic publication links
-
✓Committers with academic emails
1 of 13 committers (7.7%) from academic institutions -
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (13.0%) to scientific vocabulary
Keywords
Repository
A standalone and free application to explore genetics variations from VCF file
Basic Info
- Host: GitHub
- Owner: labsquare
- License: gpl-3.0
- Language: Python
- Default Branch: master
- Homepage: https://cutevariant.labsquare.org/
- Size: 69.2 MB
Statistics
- Stars: 107
- Watchers: 14
- Forks: 21
- Open Issues: 67
- Releases: 13
Topics
Metadata Files
README.md
Cutevariant
A standalone and free application to explore genetics variations from VCF file
Published in Bioinformatics Advanced
Documentation available on cutevariant.labsquare.org
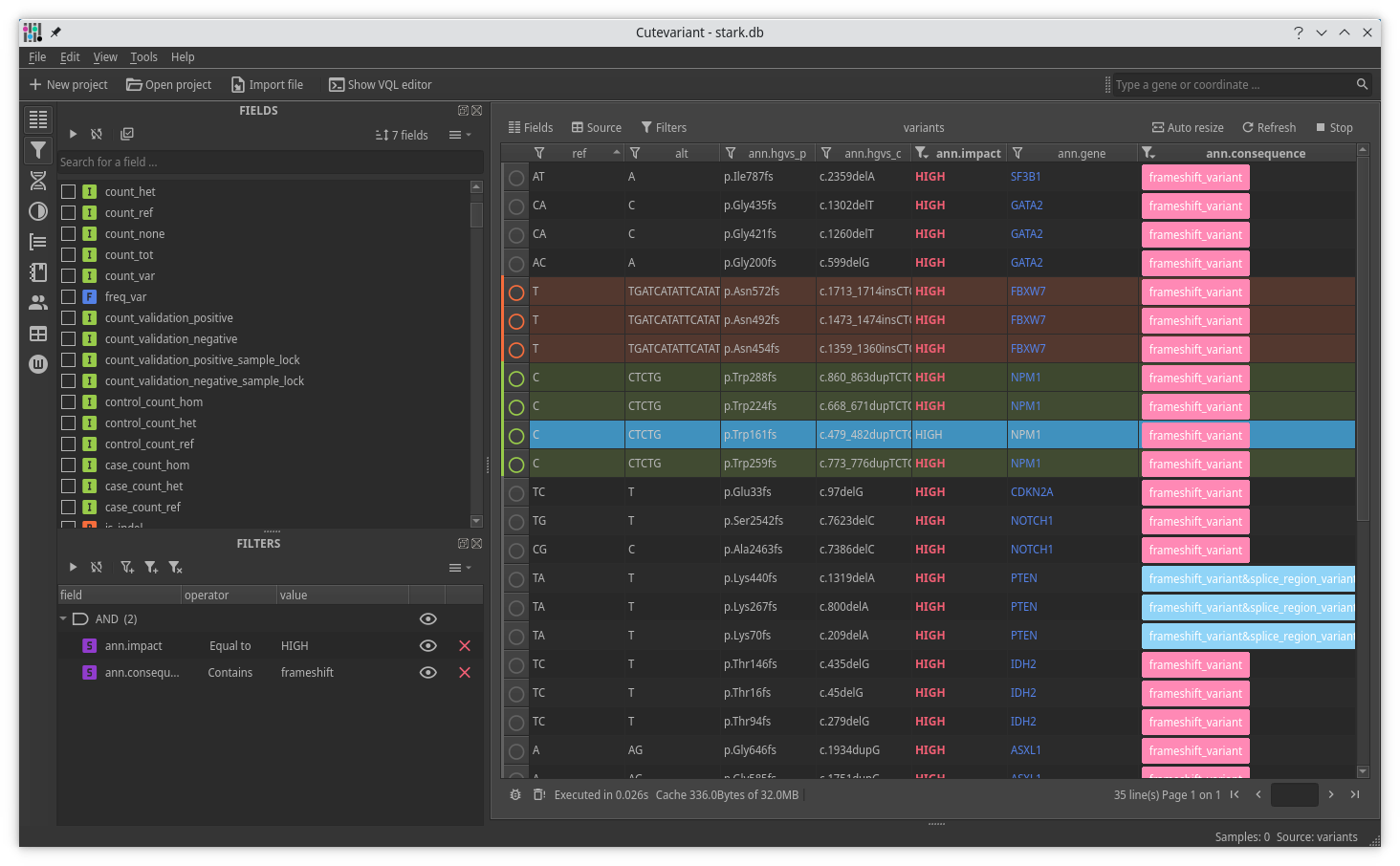
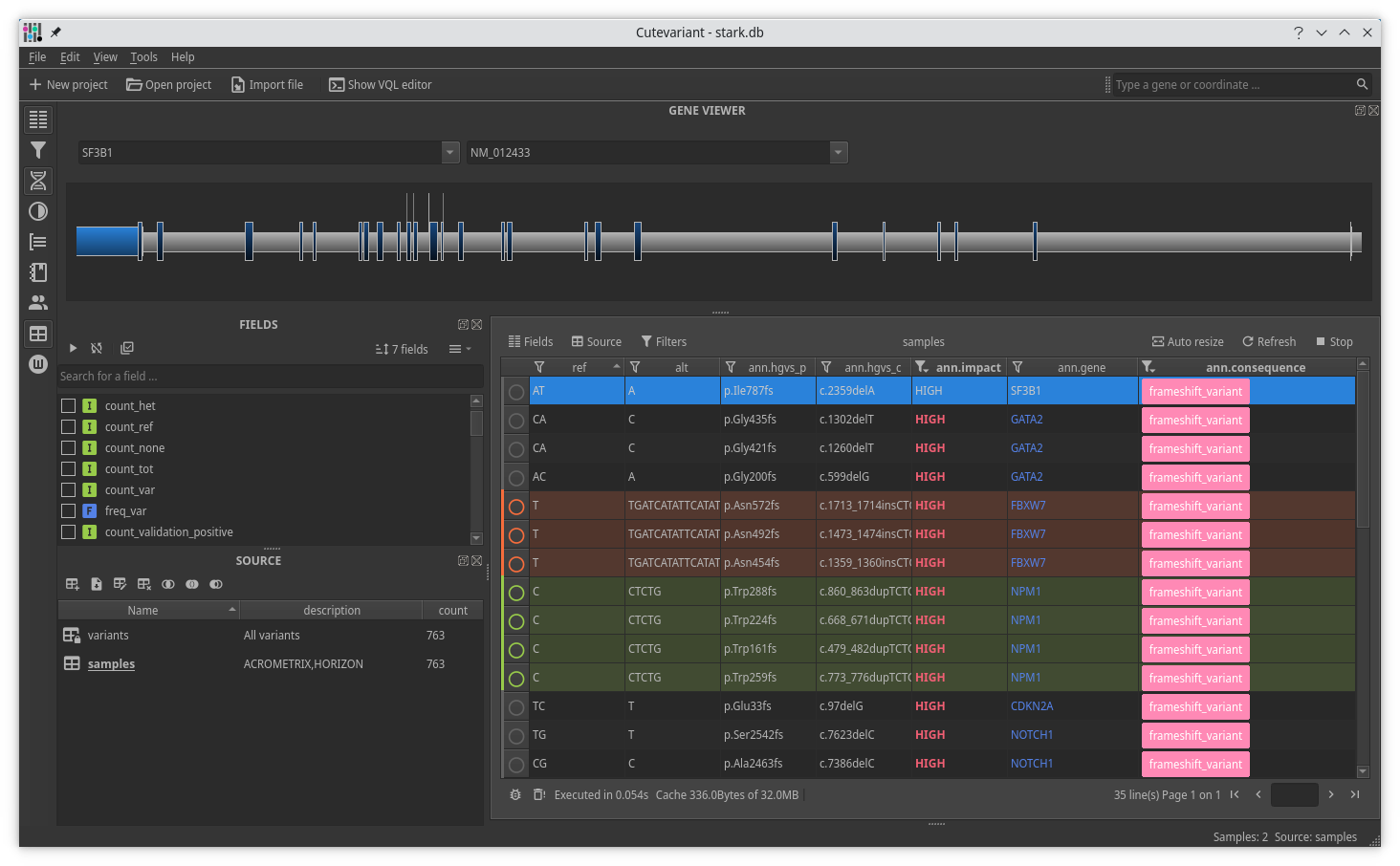
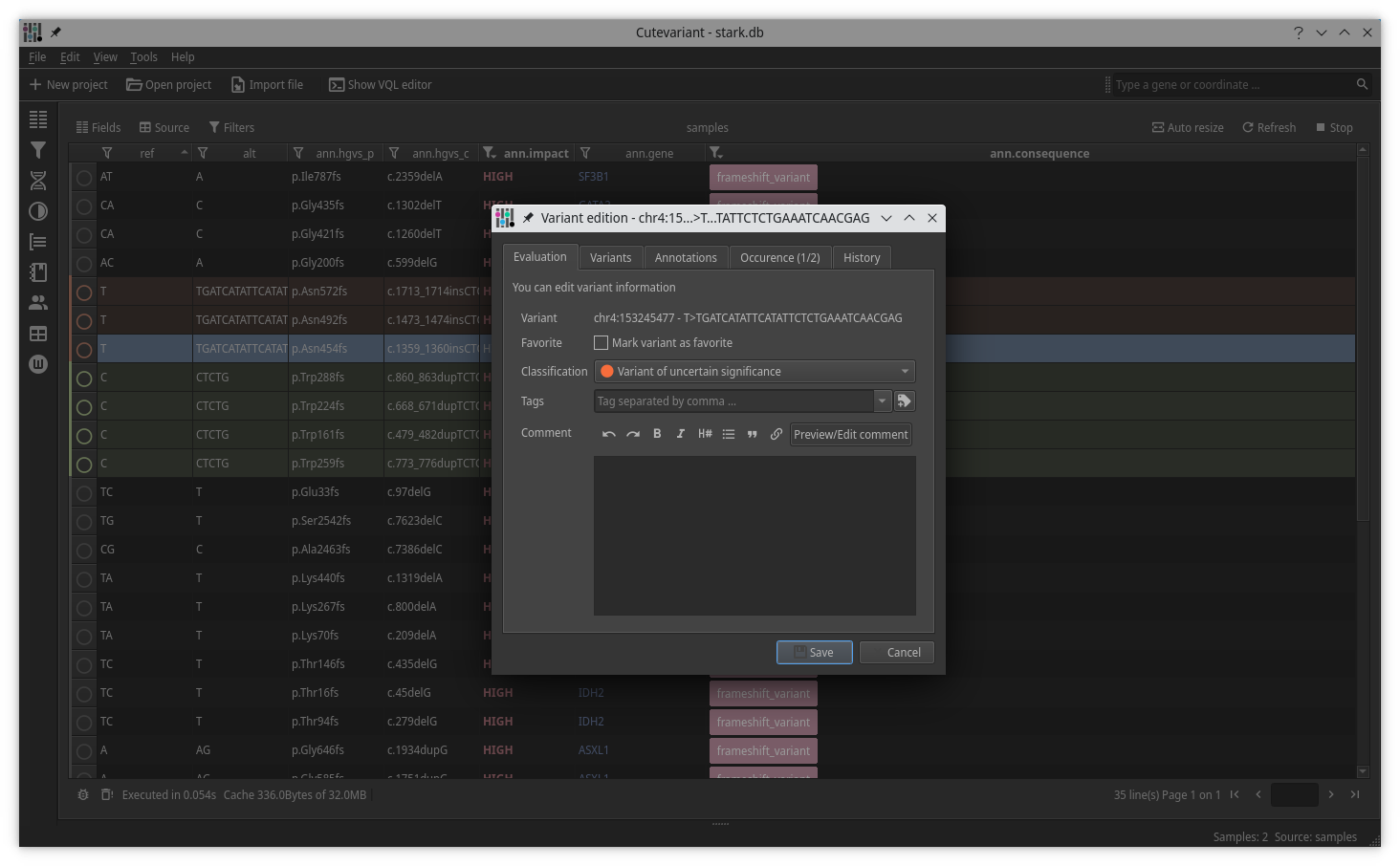
Cutevariant is a cross-plateform application dedicated to maniupulate and filter variation from annotated VCF file. When you create a project, data are imported into an sqlite database that cutevariant queries according your needs. Presently, SnpEff and VEP annotations are supported. Once your project is created, you can query variant using different gui controller or directly using the VQL language. This Domain Specific Language is specially designed for cutevariant and try to keep the same syntax than SQL for an easy use.
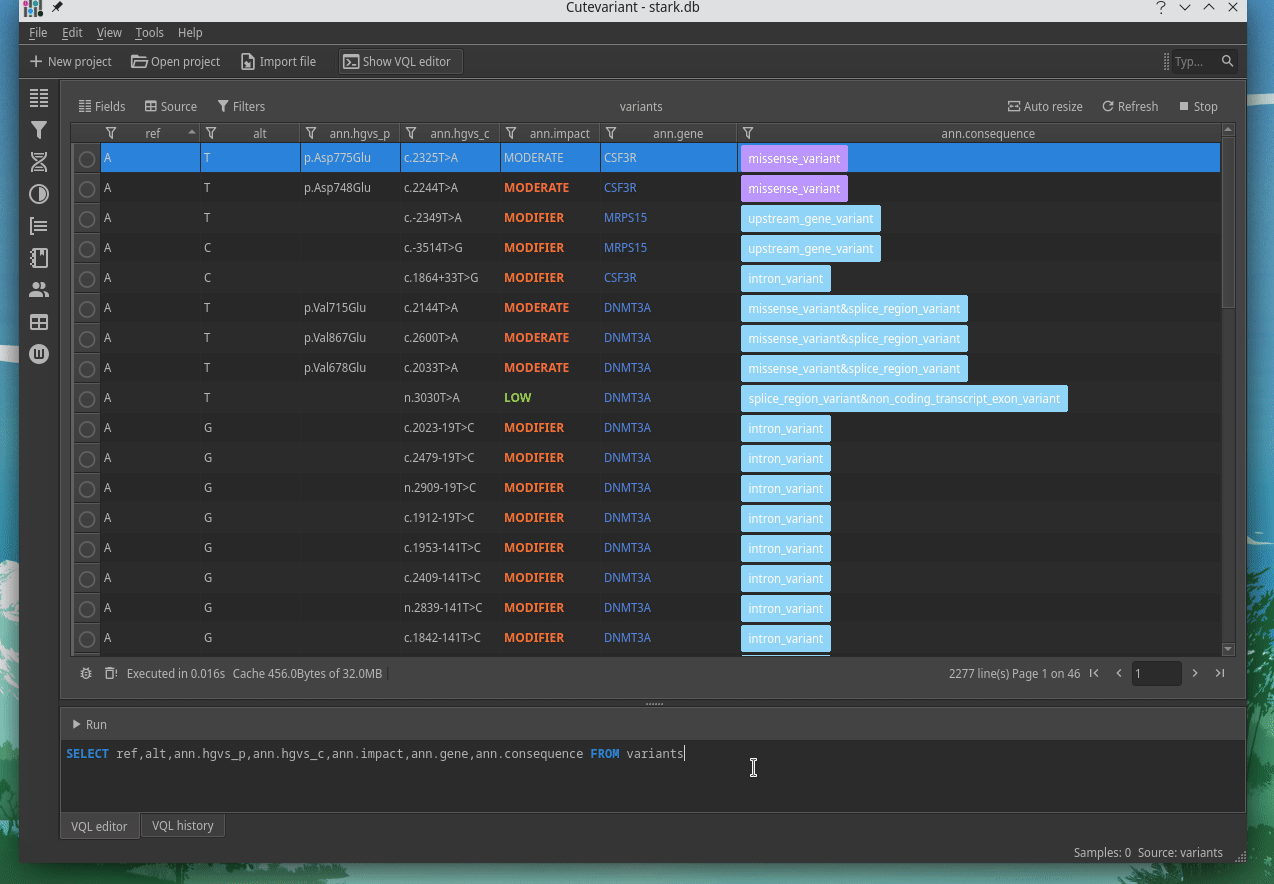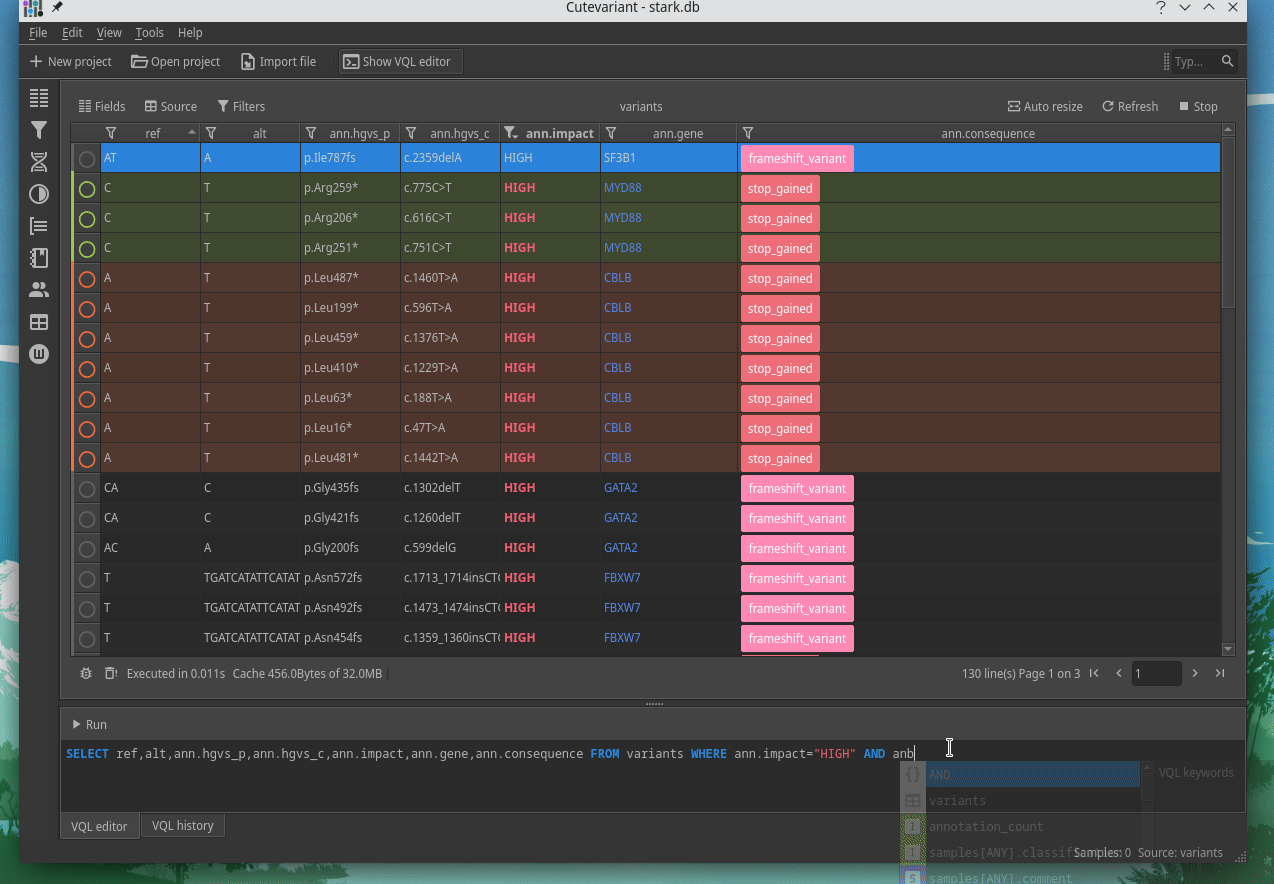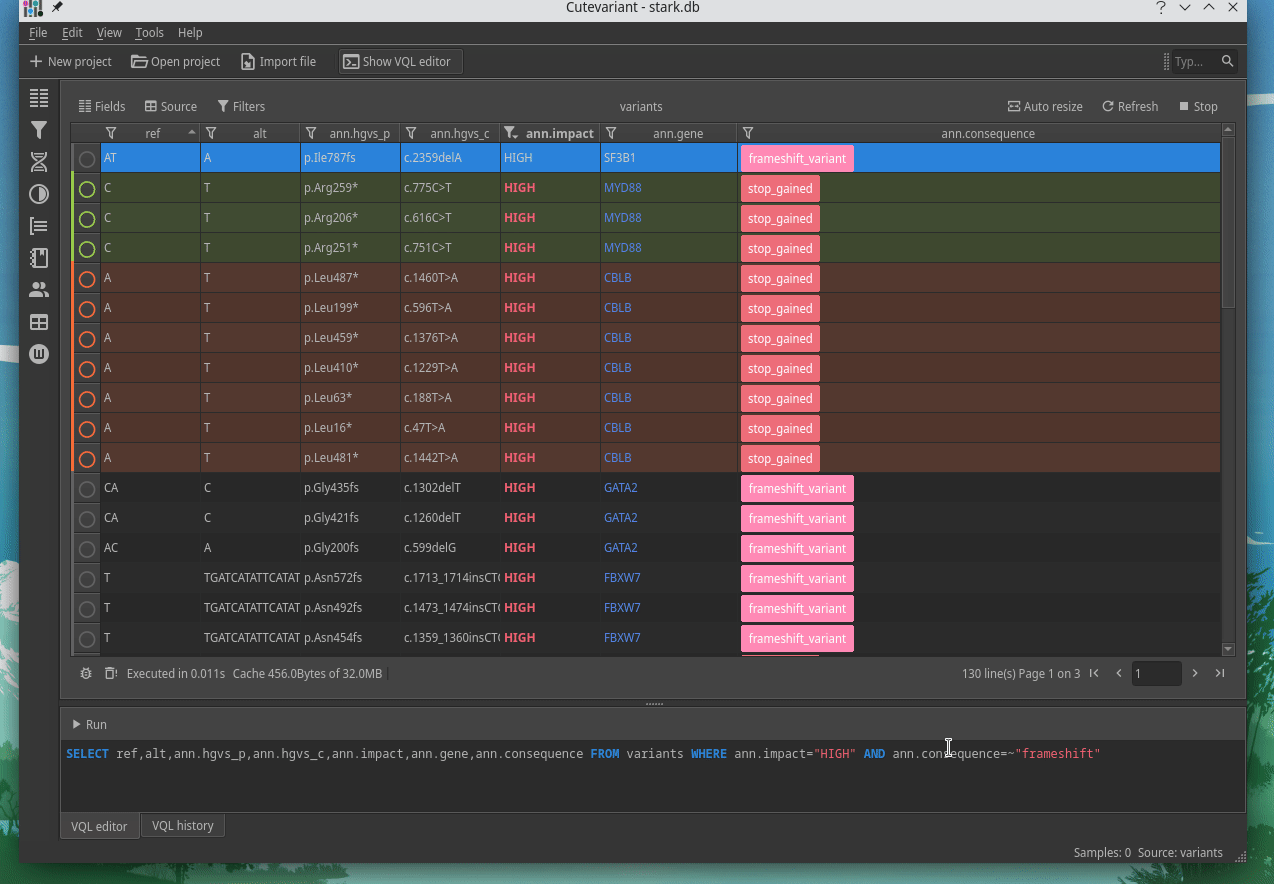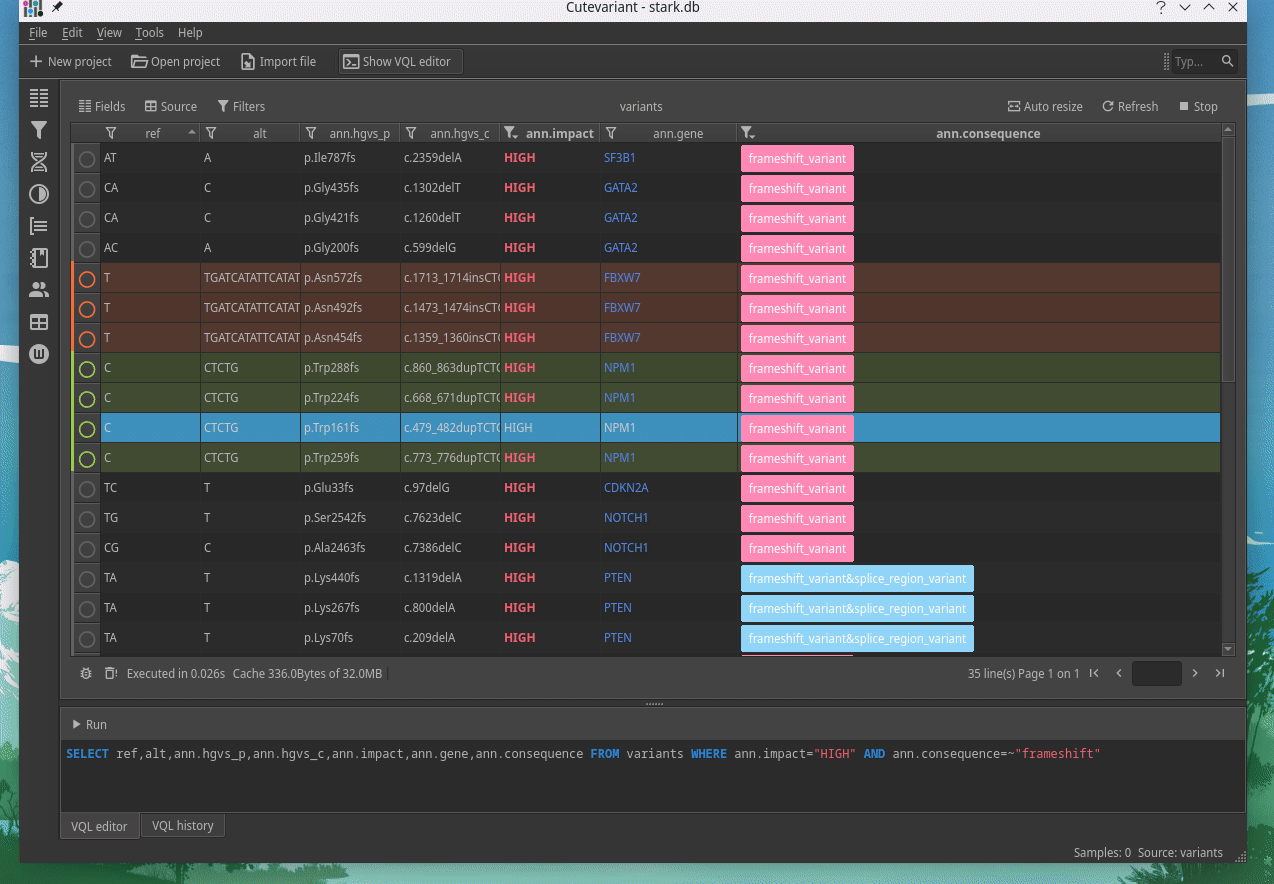
| | | |
|:-------------------------:|:-------------------------:|:-------------------------:|
| |
| |
| |
|
Installation
Windows
Standalone binary are available for windows:
- Download cutevariant 32 bit
- Download cutevariant 64 bit
Linux
If you run Linux, then you can either use PyPI or install from source.
But before you proceed to installation, make sure that running this command:
bash
sqlite3 --version
returns at least 3.32.
If not, run:
```bash
Uninstall previous versions of sqlite3 (to avoid conflicts)
sudo apt remove sqlite3
Download latest sqlite version
wget https://www.sqlite.org/2022/sqlite-autoconf-3380500.tar.gz
Extract it
tar -xvf sqlite-autoconf-3380500.tar.gz cd sqlite-autoconf-3380500 ./configure
Run make to build
make
Run make install, this will put the shared object in /usr/local/lib
sudo make install
Then add LDLIBRARYPATH to your bash profile (either ~/.zshrc, ~/.bashrc, or whatever is your favorite)
echo "export LDLIBRARYPATH=/usr/local/lib" >> ~/.zshrc
Source your shell profile so you don't have to restart it
source ~/.zshrc
Now just to be sure:
sqlite3 --version
You should see 3.38 now. If not, this means that the installation went wrong.
That's it! Now you can install cutevariant, either from PyPI or directly from source
```
PyPi
Cutevariant is avaible from Pypi :
pip install cutevariant # install
python -m cutevariant # run
From source
- Python 3.7 or newer is required
```bash
Clone repository
git clone https://github.com/labsquare/cutevariant.git cd cutevariant
Create a virtual environement
python3 -m virtualenv venv source venv/bin/activate
Install cutevariant in local mode
python -m pip install -e
Run cutevariant as module
python -m cutevariant # or make run
Run test
python -m pytest tests ```
Usages
You can follow this tutorial to familiarize yourself with cutevarant.
https://github.com/labsquare/cutevariant/wiki/Usage-examples
The VQL langage specification is available here :
https://github.com/labsquare/cutevariant/wiki/VQL-language
Contributions / Bugs
Cutevariant is a new project and all contributors are welcome
Issues
If you found a bug or have a feature request, you can report it from the Github isse trackers.
Create a plugin
Documentation to create a plugin is available here
Chat
You can join us on discord. We are speaking french right now, but we can switch to english.
Licenses
This program is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation, either version 3 of the License, or (at your option) any later version.
This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.
You should have received a copy of the GNU General Public License along with this program. If not, see https://www.gnu.org/licenses/gpl-3.0.txt.
Owner
- Name: Labsquare
- Login: labsquare
- Kind: organization
- Website: http://www.labsquare.org
- Repositories: 9
- Profile: https://github.com/labsquare
Organization for opensource genomics software
Citation (CITATION.cff)
cff-version: 1.1.0
message: "If you use this software, please cite it as below."
authors:
- family-names: Schutz
given-names: Sacha
affiliation: CHRU Brest, Hôpital Morvan, Laboratoire de Génétique Moléculaire, Brest, France
orcid: https://orcid.org/0000-0002-4563-7537
- family-names: Monod-Broca
given-names: Charles
affiliation: Univ Brest, Inserm, EFS, UMR 1078, GGB, 29200
orcid: https://orcid.org/0000-0003-4095-8099
title: labsquare/cutevariant
version: 0.3.4
date-released: 2021-06-02
GitHub Events
Total
- Watch event: 4
Last Year
- Watch event: 4
Committers
Last synced: 9 months ago
Top Committers
| Name | Commits | |
|---|---|---|
| sacha schutz | s****a@l****g | 1,567 |
| ysard | y****d | 897 |
| SteampunkIslande | c****a@n****r | 322 |
| antonylebechec | a****c@g****m | 155 |
| SamuelNicaise | i****s@g****m | 92 |
| Lucas Bourneuf | l****f@l****t | 28 |
| Valentin Klein | 5****a | 4 |
| Téo Lemane | 3****e | 1 |
| Eugene Trounev | e****v@g****m | 1 |
| sacha schutz | s****a@M****l | 1 |
| sacha | s****a@s****l | 1 |
| Felix VDM | f****n@c****r | 1 |
| Felix VANDERMEEREN | 0****1@d****d | 1 |
Committer Domains (Top 20 + Academic)
Issues and Pull Requests
Last synced: 6 months ago
All Time
- Total issues: 83
- Total pull requests: 24
- Average time to close issues: 5 months
- Average time to close pull requests: about 15 hours
- Total issue authors: 9
- Total pull request authors: 4
- Average comments per issue: 1.01
- Average comments per pull request: 1.67
- Merged pull requests: 19
- Bot issues: 0
- Bot pull requests: 0
Past Year
- Issues: 1
- Pull requests: 0
- Average time to close issues: N/A
- Average time to close pull requests: N/A
- Issue authors: 1
- Pull request authors: 0
- Average comments per issue: 0.0
- Average comments per pull request: 0
- Merged pull requests: 0
- Bot issues: 0
- Bot pull requests: 0
Top Authors
Issue Authors
- antonylebechec (40)
- dridk (19)
- SamuelNicaise (17)
- splaisan (2)
- iaacornus (1)
- ccarco (1)
- SteampunkIslande (1)
- bioinfo-chru-strasbourg (1)
- smanilov (1)
Pull Request Authors
- antonylebechec (12)
- SamuelNicaise (6)
- ikit (3)
- dridk (3)
Top Labels
Issue Labels
Pull Request Labels
Packages
- Total packages: 1
-
Total downloads:
- pypi 66 last-month
- Total dependent packages: 0
- Total dependent repositories: 1
- Total versions: 36
- Total maintainers: 2
pypi.org: cutevariant
GUI to visualize and process variant data
- Homepage: https://github.com/labsquare/cutevariant
- Documentation: https://cutevariant.readthedocs.io/
- License: GNU General Public License v3 or later (GPLv3+)
-
Latest release: 0.4.5
published almost 3 years ago
Rankings
Dependencies
- actions/checkout v2 composite
- actions/setup-python v2 composite
- codecov/codecov-action v2 composite