vuegen
VueGen automates the creation of reports from bioinformatics outputs, supporting formats like PDF, HTML, DOCX, ODT, PPTX, Reveal.js, Jupyter notebooks, and Streamlit web applications. Users simply provide a directory with output files and VueGen compiles them into a structured report.
Science Score: 67.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
✓CITATION.cff file
Found CITATION.cff file -
✓codemeta.json file
Found codemeta.json file -
✓.zenodo.json file
Found .zenodo.json file -
✓DOI references
Found 11 DOI reference(s) in README -
✓Academic publication links
Links to: zenodo.org -
○Academic email domains
-
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (11.3%) to scientific vocabulary
Keywords
Repository
VueGen automates the creation of reports from bioinformatics outputs, supporting formats like PDF, HTML, DOCX, ODT, PPTX, Reveal.js, Jupyter notebooks, and Streamlit web applications. Users simply provide a directory with output files and VueGen compiles them into a structured report.
Basic Info
- Host: GitHub
- Owner: Multiomics-Analytics-Group
- License: mit
- Language: Python
- Default Branch: main
- Homepage: https://vuegen.readthedocs.io
- Size: 84.3 MB
Statistics
- Stars: 26
- Watchers: 2
- Forks: 0
- Open Issues: 28
- Releases: 12
Topics
Metadata Files
README.md
VueGen is a Python package that automates the creation of scientific reports.
| Information | Links |
| :-------------------- | :-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Package | 






|
| Examples |





Table of contents:
- About the project
- Installation
- Execution
- GUI
- Case studies
- Web application deployment
- License
- Contributing
- Credits and acknowledgements
- Citation
- Contact and feedback
About the project
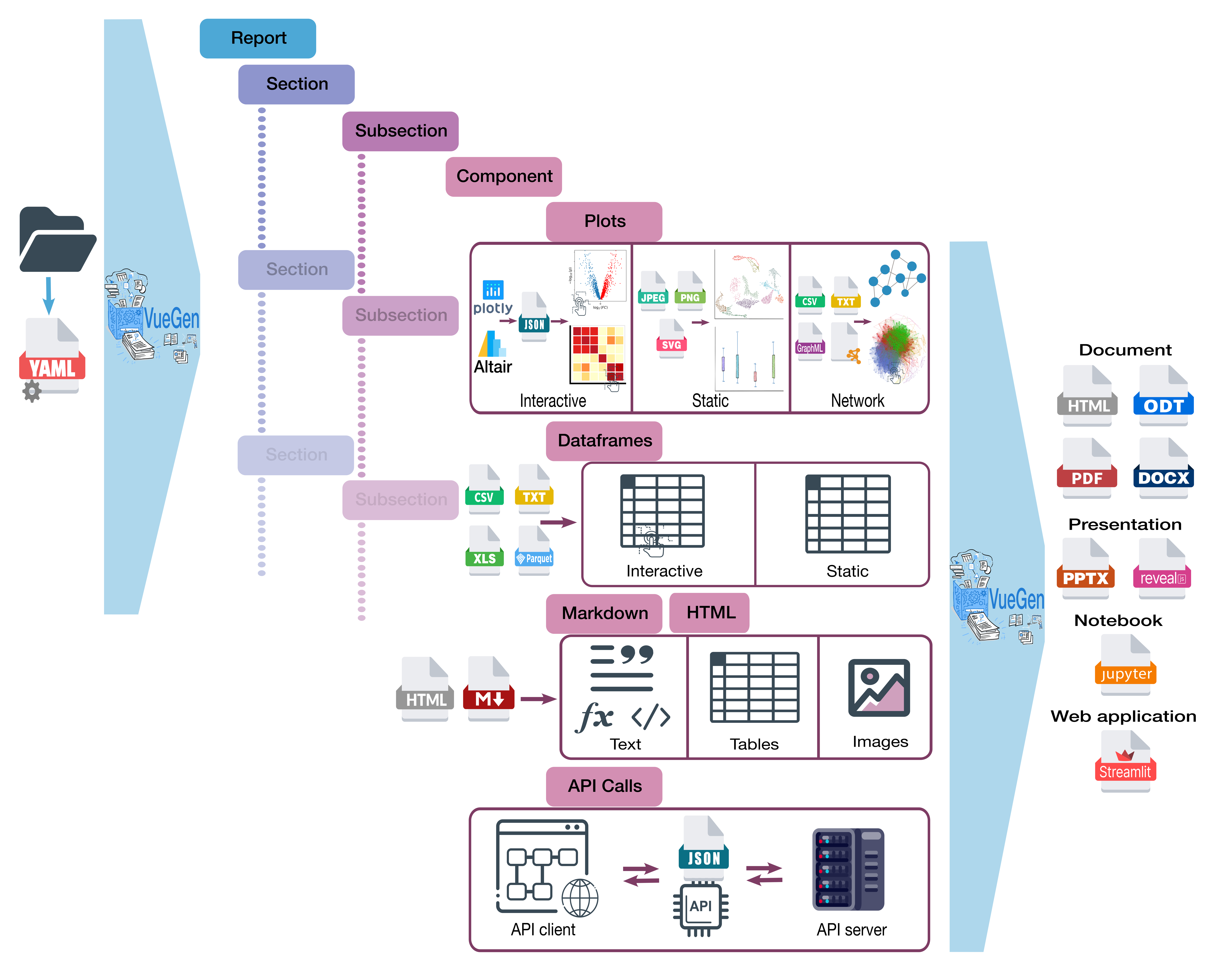
VueGen is a tool that automates the creation of reports from bioinformatics outputs, allowing researchers with minimal coding experience to communicate their results effectively. With VueGen, users can produce reports by simply specifying a directory containing output files, such as plots, tables, networks, Markdown text, HTML components, and API calls, along with the report format. Supported formats include documents (PDF, HTML, DOCX, ODT), presentations (PPTX, Reveal.js), Jupyter notebooks, and Streamlit web applications.
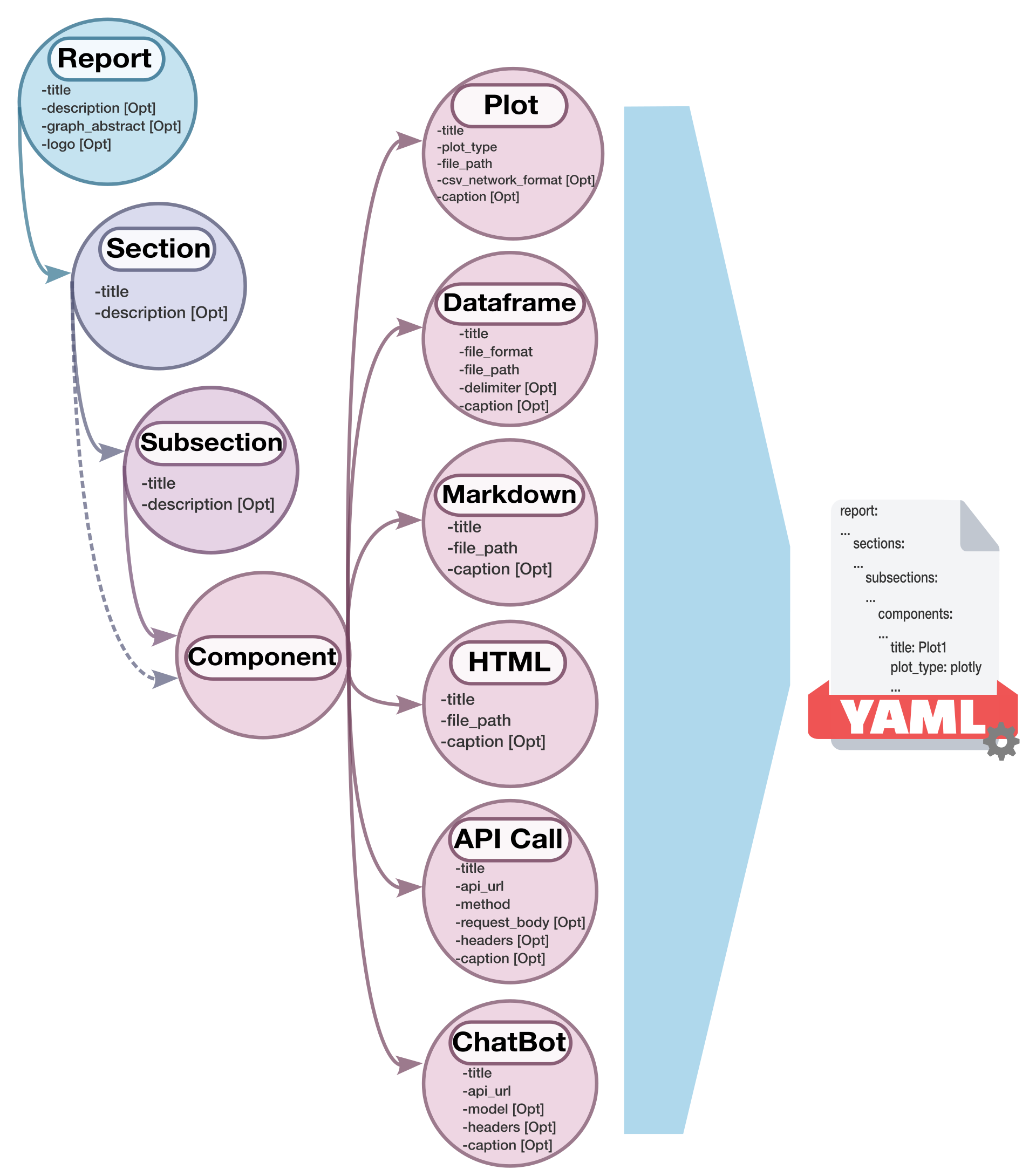
A YAML configuration file is generated from the directory to define the structure of the report. Users can customize the report by modifying the configuration file, or they can create their own configuration file instead of passing a directory as input. The configuration file specifies the structure of the report, including sections, subsections, and various components such as plots, dataframes, markdown, html, and API calls.
An overview of the VueGen workflow is shown in the figure below:
We created a schema diagram to illustrates the structure of the configuration file and the relationships between its elements:
Also, the class diagram for the project's current version is presented below to show the architecture and relationships between classes:
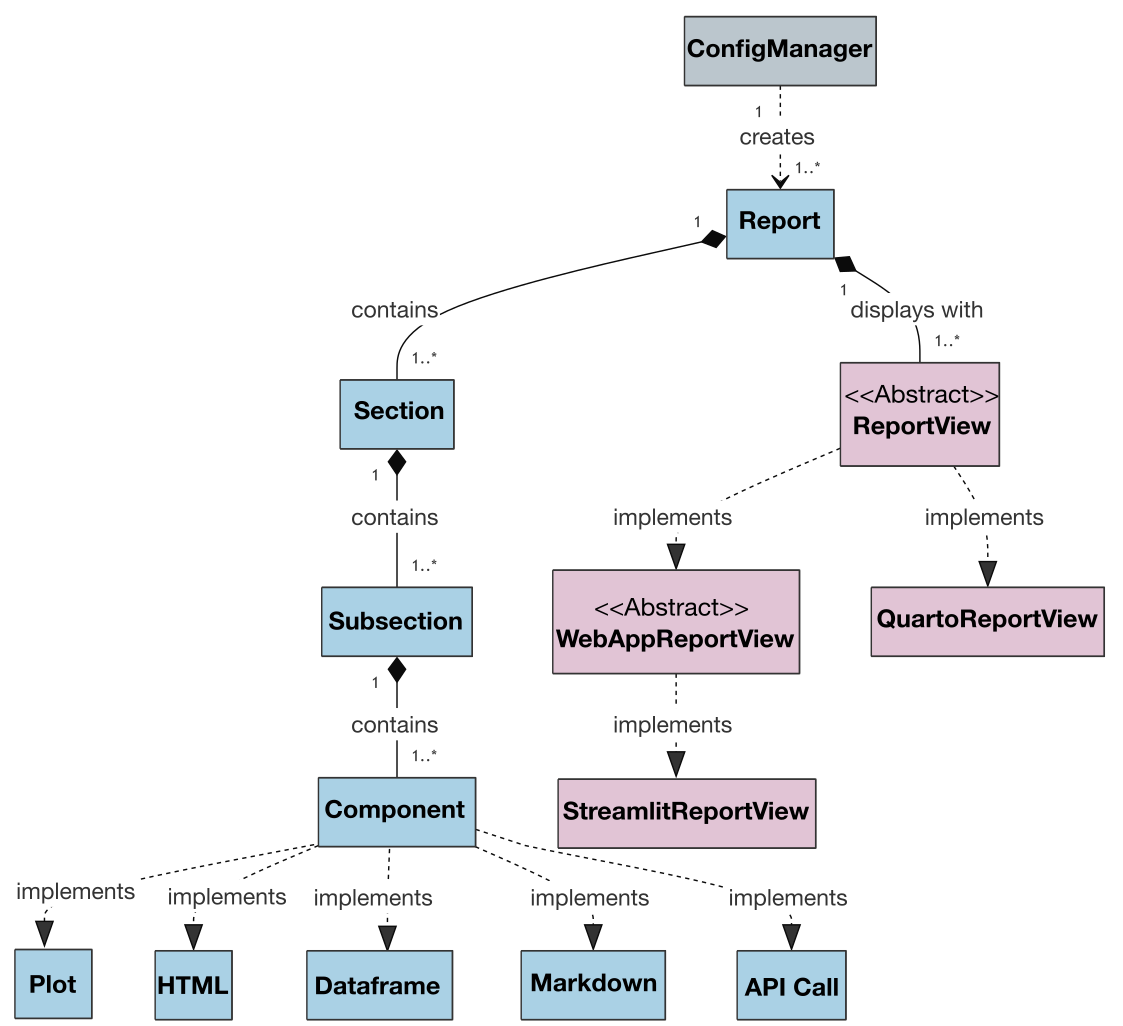
An extended version of the class diagram with attributes and methods is available here.
The VueGen documentation is available at vuegen.readthedocs.io, where you can find detailed information of the package’s classes and functions, installation and execution instructions, and case studies to demonstrate its functionality.
Installation
[!TIP] It is recommended to install VueGen inside a virtual environment to manage depenendencies and avoid conflicts with existing packages. You can use the virtual environment manager of your choice, such as
poetry,conda, orpipenv.
Pip
VueGen is available on PyPI and can be installed using pip:
bash
pip install vuegen
You can also install the package for development by cloning this repository and running the following command:
[!WARNING] We assume you are in the root directory of the cloned repository when running this command. Otherwise, you need to specify the path to the
vuegendirectory.
bash
pip install -e .
Conda
VueGen is also available on Bioconda and can be installed using conda:
bash
conda install -c bioconda -c conda-forge vuegen
Dependencies
VueGen uses Quarto to generate various report types. The pip insallation includes quarto using the quarto-cli Python library. To test if quarto is installed in your computer, run the following command:
bash
quarto check
[!TIP] If quarto is not installed, you can download the command-line interface from the Quarto website for your operating system.
For PDF reports, you need to have a LaTeX distribution installed. This can be done with quarto using the following command:
bash
quarto install tinytex
[!TIP] Also, you can add the
--quarto_checksargument to the VueGen command to check and install the required dependencies automatically.
Docker
If you prefer not to install VueGen on your system, a pre-configured Docker container is available. It includes all dependencies, ensuring a fully reproducible execution environment. See the Execution section for details on running VueGen with Docker. The official Docker images are available at quay.io/dtubiosustaindsp/vuegen. The Dockerfiles to build the images are available here.
Nextflow and nf-core
VueGen is also available as a nf-core module, customised for compatibility with the Nextflow environment. This module is designed to automate report generation from outputs produced by other modules, subworkflows, or pipelines. Asumming that you have nextflow and nf-core installed, you can use the following command to install the nf-core module:
bash
nf-core modules install vuegen
[!NOTE] You can read the offical documentation for the nf-core module here. Also, the source code and additional details are available in the nf-VueGen repository.
Execution
[!IMPORTANT] Here we use the
Earth_microbiome_vuegen_demo_notebookdirectory and theEarth_microbiome_vuegen_demo_notebook.yamlconfiguration file as examples, which are available in thedocs/example_dataanddocs/example_config_filesfolders, respectively. Make sure to clone the VueGen's GitHub reposiotry to access these contents, or use your own directory and configuration file.
Run VueGen using a directory with the following command:
bash
vuegen --directory docs/example_data/Earth_microbiome_vuegen_demo_notebook --report_type streamlit
[!NOTE] By default, the
streamlit_autorunargument is set to False, but you can use it in case you want to automatically run the streamlit app. You can also specify the output directory with the--output_directoryargumument, which defaults to the current working directory. See all available arguments with the--helpoption.
Folder structure
Your input directory should follow a nested folder structure, where first-level folders are treated as sections and second-level folders as subsections, containing the components (plots, tables, networks, Markdown text, and HTML files). If the component files are in the first-level folders, an overview subsection will be created automatically.
Here is an example layout:
report_folder/
├── section1/
│ ├── table1.tsv
│ └── subsection1/
│ ├── table2.csv
│ ├── image1.png
│ └── chart.json
├── section2/
│ ├── image2.jpg
│ ├── subsection1/
│ │ ├── summary_table.xls
│ │ └── network_plot.graphml
│ └── subsection2/
│ ├── report.html
│ └── summary.md
The titles for sections, subsections, and components are extracted from the corresponding folder and file names, and afterward, users can add descriptions, captions, and other details to the configuration file. Component types are inferred from the file extensions and names. The order of sections, subsections, and components can be defined using numerical suffixes in folder and file names.
Configuration file
It's also possible to provide a configuration file instead of a directory:
bash
vuegen --config docs/example_config_files/Earth_microbiome_vuegen_demo_notebook.yaml --report_type streamlit
If a configuration file is given, users can specify titles and descriptions for sections and subsections, as well as component paths and required attributes, such as file format and delimiter for dataframes, plot types, and other details.
The component paths in the configuration file can be absolute or relative to the execution directory. In the examples, we assume that the working directory is the docs folder, so the paths are relative to it. If you run VueGen from another directory, you need to adjust the paths accordingly.
The current report types supported by VueGen are:
- Streamlit
- HTML
- DOCX
- ODT
- Reveal.js
- PPTX
- Jupyter
Running VueGen with Docker
Instead of installing VueGen locally, you can run it directly from a Docker container with the following command:
bash
docker run --rm \
-v "$(pwd)/docs/example_data/Earth_microbiome_vuegen_demo_notebook:/home/appuser/Earth_microbiome_vuegen_demo_notebook" \
-v "$(pwd)/output_docker:/home/appuser/streamlit_report" \
quay.io/dtu_biosustain_dsp/vuegen:v0.3.2-docker --directory /home/appuser/Earth_microbiome_vuegen_demo_notebook --report_type streamlit
Running VueGen with Nextflow and nf-core
To run VueGen as a nf-core module, you should create a Nextflow pipeline and include the VueGen module in your workflow. Here is a main.nf example:
```groovy
!/usr/bin/env nextflow
include { VUEGEN } from './modules/nf-core/vuegen/'
workflow { // Create a channel for the report type reporttypech = Channel.value(params.report_type)
// Handle configuration file and directory inputs
if (params.config) {
file_ch = Channel.fromPath(params.config)
input_type_ch = Channel.value('config')
output_ch = VUEGEN(input_type_ch, file_ch, report_type_ch)
} else if (params.directory) {
dir_ch = Channel.fromPath(params.directory, type: 'dir', followLinks: true)
input_type_ch = Channel.value('directory')
output_ch = VUEGEN(input_type_ch, dir_ch, report_type_ch)
}
} ```
You can run the pipeline with the following command:
bash
nextflow run main.nf --directory docs/example_data/Basic_example_vuegen_demo_notebook --report_type html
[!NOTE] You can read the offical documentation for the nf-core module here. Also, the source code and additional details are available in the nf-VueGen repository.
GUI
We have a simple GUI for VueGen that can be run locally or through a standalone executable.
Local GUI
To use the local GUI, you should clone this repository and install the required dependencies. You can do this by running the following command in the root directory of the cloned repository:
bash
pip install '.[gui]'
Then, you should move to the gui folder and execute the app.py Python file:
bash
python app.py
Bundled GUI
The bundle GUI with the VueGen package is available under the
latest releases.
You will need to unzip the file and run vuegen_gui in the unpacked main folder.
Most dependencies are included into the bundle using PyInstaller.
Streamlit works out of the box as a purely Python based package. For the rest of report types you
will have to have a Python 3.12 installation with the jupyter package installed, as quarto needs to start
a kernel for execution. This is also true if you install quarto globally on your machine.
[!TIP] It is advisable to create a virtual environment to manage depenendencies and avoid conflicts with existing packages. You can use the virtual environment manager of your choice, such as
poetry,conda, orpipenv. We recommend using miniforge to install Python and thecondapackage manager.
We assume you have installed the miniforge distribution for your machine (MacOS with arm64/ apple silicon
or x8664/ intel or Windows x8664). Also, download the
latest vuegen_gui bundle
from the releases page according to your operating system.
You can create a new conda environment with Python 3.12 and the jupyter and vuegen package:
bash
conda create -n vuegen_gui -c conda-forge -c bioconda python=3.12 jupyter vuegen
The exact Python version used for the release is specified on the releases page as we observed that sometimes the exact Python version is required to run the GUI.
[!WARNING] If you have errors with the
vuegenpackage, you can install it separately using pip, as explained in the installation section.
conda create -n vuegen_gui -c conda-forge -c bioconda python=3.12 jupyter conda activate vuegen_gui pip install vuegen
Then, activate the environment:
bash
conda activate vuegen_gui
Now, you can list all conda environments to find the location of the vuegen_gui environment:
bash
conda info -e
On MacOS, you need to add a bin to the path:
bash
/Users/user/miniforge3/envs/vuegen_gui/bin
On Windows, you can use the path as displayed by conda info -e:
bash
C:\Users\user\miniforge3\envs\vuegen_gui
[!NOTE] On Windows a base installation of miniforge with
jupytermight work because the app can see your entire Path, which is not the case on MacOS.
More information regarding the app and builds can be found in the GUI README.
Case studies
VueGen’s functionality is demonstrated through various case studies:
1. Predefined Directory
This introductory case study uses a predefined directory with plots, dataframes, Markdown, and HTML components. Users can generate reports in different formats and modify the configuration file to customize the report structure.
[!NOTE] The configuration file is available in the
docs/example_config_filesfolder, and the directory with example data is in thedocs/example_datafolder.
2. Earth Microbiome Project Data
This advanced case study demonstrates the application of VueGen in a real-world scenario using data from the Earth Microbiome Project (EMP). The EMP is an initiative to characterize global microbial taxonomic and functional diversity. The notebook process the EMP data, create plots, dataframes, and other components, and organize outputs within a directory to produce reports. Report content and structure can be adapted by modifying the configuration file. Each report consists of sections on exploratory data analysis, metagenomics, and network analysis.
[!NOTE] The EMP case study is available online as HTML and Streamlit reports. The configuration file is available in the
docs/example_config_filesfolder, and the directory with example data is in thedocs/example_datafolder.
3. APICall Component
This case study focuses on the APICall component, which enables interaction with external APIs by using HTTP methods such as GET and POST. The retrieved data is displayed in the report, allowing users to integrate external data sources into their anlyses. This component is restricted to Streamlit reports.
[!NOTE] A configuration file example for the apicall component is available in the
docs/example_config_filesfolder.
4. ChatBot Component
This case study highlights VueGen’s capability to embed a chatbot component into a report subsection, enabling interactive conversations inside the report. This component is streamlit-specific and is not available for other report types.
Two API modes are supported:
- Ollama-style streaming chat completion:
If a
modelparameter is specified in the config file, VueGen assumes the chatbot is using Ollama’s /api/chat endpoint. Messages are handled as chat history, and the assistant responses are streamed in real time for a smooth and responsive experience. This mode supports LLMs such asllama3,deepsek, ormistral.
[!TIP] See Ollama’s website for more details.
- Standard prompt-response API:
If no
modelis provided, VueGen uses a simpler prompt-response flow. A single prompt is sent to an endpoint, and a structured JSON object is expected in return. Currently, the response can include:text: the main textual replylinks: a list of source URLs (optional)HTML content: an HTML snippet with a Pyvis network visualization (optional)
This response structure is currently customized for an internal knowledge graph assistant, but VueGen is being actively developed to support more flexible and general-purpose response formats in future releases.
[!NOTE] A configuration file example for the chatbot component is available in the
docs/example_config_filesfolder.
Web application deployment
Once a Streamlit report is generated, it can be deployed as a web application to make it accessible online. There are multiple ways to achieve this:
Streamlit Community Cloud
Deploy your report easily using Streamlit Cloud, as demonstrated in the EMP VueGen Demo. The process involves moving the necessary scripts, data, and a requirements.txt file into a GitHub repository. Then, the app can be deployed via the Streamlit Cloud interface. The deployment example is available in the streamlit-report-example branch.
Standalone Executables
Convert your Streamlit application into a desktop app by packaging it as an executable file for different operating systems. A detailed explanation of this process can be found in this Streamlit forum post.
Stlite
Run Streamlit apps directly in the browser with stlite, a WebAssembly port of Streamlit powered by Pyodide, eliminating the need for a server. It also allows packaging apps as standalone desktop executables using stlite desktop.
These options provide flexibility depending on whether the goal is online accessibility, lightweight execution, or local application distribution.
License
The code in this repository is licensed under the MIT License, allowing you to use, modify, and distribute it freely as long as you include the original copyright and license notice.
The documentation and other creative content are licensed under the Creative Commons Attribution 4.0 International (CC BY 4.0) License, meaning you are free to share and adapt it with proper attribution.
Full details for both licenses can be found in the LICENSE file.
Contributing
VueGen is an open-source project, and we welcome contributions of all kinds via GitHub issues and pull requests. You can report bugs, suggest improvements, propose new features, or implement changes. Please follow the guidelines in the CONTRIBUTING file to ensure that your contribution is easily integrated into the project.
Credits and acknowledgements
- VueGen was developed by the Multiomics Network Analytics Group (MoNA) at the Novo Nordisk Foundation Center for Biosustainability (DTU Biosustain).
- VueGen relies on the work of numerous open-source projects like Streamlit, Quarto, and others. A big thank you to their authors for making this possible!
- The vuegen logo was designed based on an image created by Scriberia for The Turing Way Community, which is shared under a CC-BY licence. The original image can be found at Zenodo.
Citation
If you use VueGen in your research or publications, please cite it as follows:
APA:
Ayala-Ruano, S., Webel, H., & Santos, A. (2025). VueGen: Automating the generation of scientific reports. Bioinformatics Advances, vbaf149. https://doi.org/10.1093/bioadv/vbaf149
BibTeX:
bibtex
@article{10.1093/bioadv/vbaf149,
author = {Ayala-Ruano, Sebastian and Webel, Henry and Santos, Alberto},
title = {VueGen: Automating the generation of scientific reports},
journal = {Bioinformatics Advances},
pages = {vbaf149},
year = {2025},
month = {06},
issn = {2635-0041},
doi = {10.1093/bioadv/vbaf149},
url = {https://doi.org/10.1093/bioadv/vbaf149},
eprint = {https://academic.oup.com/bioinformaticsadvances/advance-article-pdf/doi/
10.1093/bioadv/vbaf149/63568410/vbaf149.pdf},
}
Contact and feedback
We appreciate your feedback! If you have any comments, suggestions, or run into issues while using VueGen, feel free to open an issue in this repository. Your input helps us make VueGen better for everyone.
Owner
- Name: Multi-omics Network Analytics Group
- Login: Multiomics-Analytics-Group
- Kind: organization
- Email: albsad@biosustain.dtu.dk
- Location: Denmark
- Repositories: 1
- Profile: https://github.com/Multiomics-Analytics-Group
Citation (CITATION.cff)
cff-version: 1.2.0
message: "If you use VueGen in your research, please cite it as below."
title: "VueGen: Automating the generation of scientific reports"
version: 1.0.0
authors:
- family-names: "Ayala-Ruano"
given-names: "Sebastian"
orcid: "http://orcid.org/0000-0001-9756-6745"
- family-names: "Webel"
given-names: "Henry"
orcid: "http://orcid.org/0000-0001-8833-7617"
- family-names: "Santos"
given-names: "Alberto"
orcid: "http://orcid.org/0000-0002-9163-7730"
doi: "10.1101/2025.03.05.641152"
date-released: "2025-03-05"
url: "https://github.com/YOUR_GITHUB_USERNAME/YOUR_REPO_NAME"
preferred-citation:
type: article
authors:
- family-names: "Ayala-Ruano"
given-names: "Sebastian"
orcid: "http://orcid.org/0000-0001-9756-6745"
- family-names: "Webel"
given-names: "Henry"
orcid: "http://orcid.org/0000-0001-8833-7617"
- family-names: "Santos"
given-names: "Alberto"
orcid: "http://orcid.org/0000-0002-9163-7730"
title: "VueGen: Automating the generation of scientific reports"
journal: "Bioinformatics Advances"
year: 2025
doi: "10.1093/bioadv/vbaf149"
url: "https://doi.org/10.1093/bioadv/vbaf149"
GitHub Events
Total
- Create event: 64
- Release event: 11
- Issues event: 72
- Watch event: 19
- Delete event: 53
- Issue comment event: 68
- Public event: 1
- Push event: 346
- Pull request review comment event: 58
- Pull request review event: 81
- Pull request event: 91
- Fork event: 1
Last Year
- Create event: 64
- Release event: 11
- Issues event: 72
- Watch event: 19
- Delete event: 53
- Issue comment event: 68
- Public event: 1
- Push event: 346
- Pull request review comment event: 58
- Pull request review event: 81
- Pull request event: 91
- Fork event: 1
Issues and Pull Requests
Last synced: 6 months ago
All Time
- Total issues: 89
- Total pull requests: 106
- Average time to close issues: 17 days
- Average time to close pull requests: 5 days
- Total issue authors: 2
- Total pull request authors: 2
- Average comments per issue: 0.27
- Average comments per pull request: 0.89
- Merged pull requests: 88
- Bot issues: 0
- Bot pull requests: 0
Past Year
- Issues: 89
- Pull requests: 106
- Average time to close issues: 17 days
- Average time to close pull requests: 5 days
- Issue authors: 2
- Pull request authors: 2
- Average comments per issue: 0.27
- Average comments per pull request: 0.89
- Merged pull requests: 88
- Bot issues: 0
- Bot pull requests: 0
Top Authors
Issue Authors
- sayalaruano (56)
- enryH (33)
Pull Request Authors
- enryH (64)
- sayalaruano (41)
