gget
🧬 gget enables efficient querying of genomic reference databases
Science Score: 67.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
✓CITATION.cff file
Found CITATION.cff file -
✓codemeta.json file
Found codemeta.json file -
✓.zenodo.json file
Found .zenodo.json file -
✓DOI references
Found 4 DOI reference(s) in README -
○Academic publication links
-
✓Committers with academic emails
3 of 18 committers (16.7%) from academic institutions -
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (14.8%) to scientific vocabulary
Keywords
Repository
🧬 gget enables efficient querying of genomic reference databases
Basic Info
- Host: GitHub
- Owner: pachterlab
- License: bsd-2-clause
- Language: Python
- Default Branch: main
- Homepage: https://gget.bio
- Size: 330 MB
Statistics
- Stars: 1,049
- Watchers: 8
- Forks: 77
- Open Issues: 20
- Releases: 36
Topics
Metadata Files
README.md
gget
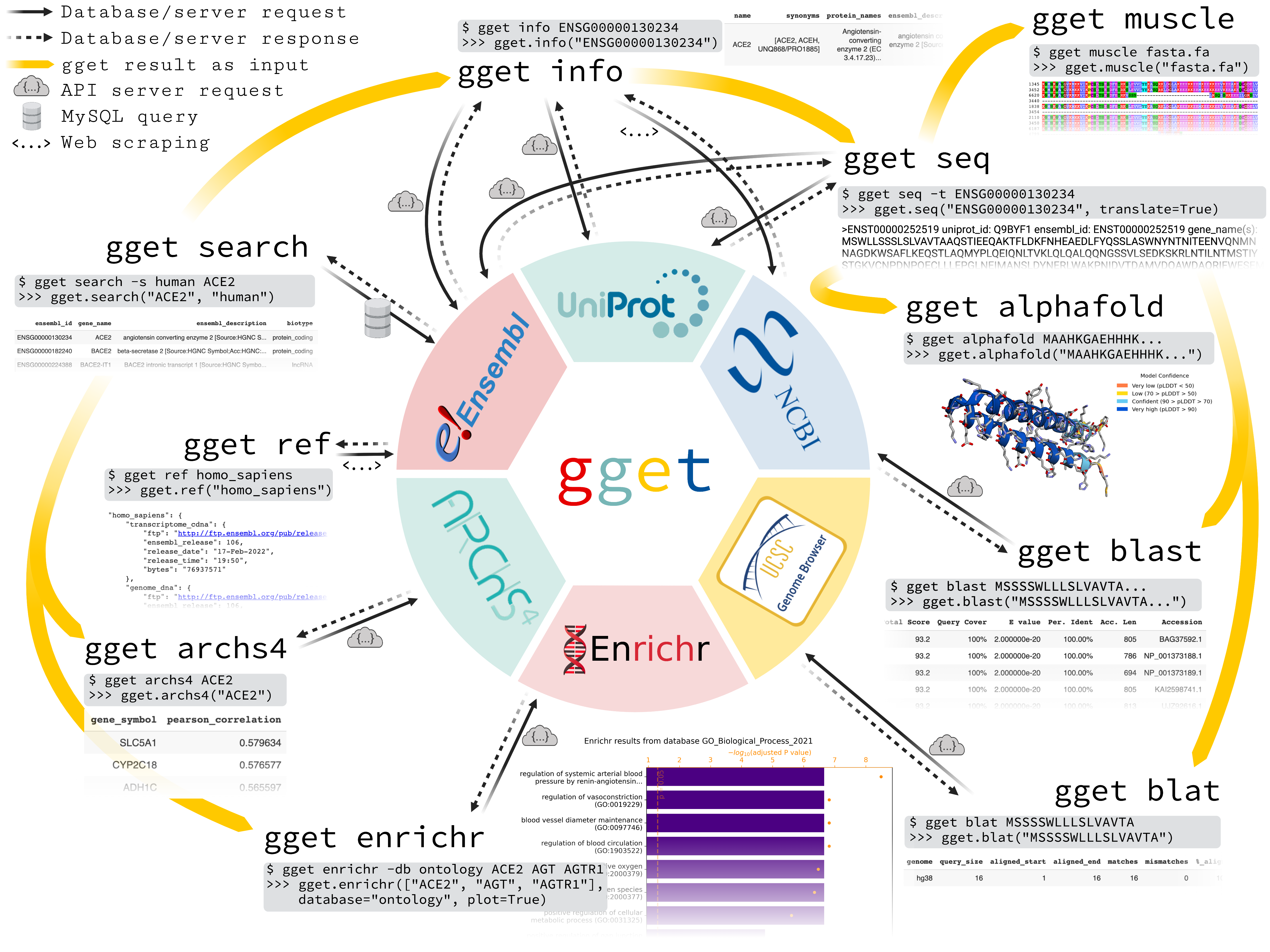
gget is a free, open-source command-line tool and Python package that enables efficient querying of genomic databases. gget consists of a collection of separate but interoperable modules, each designed to facilitate one type of database querying in a single line of code.
If you use gget in a publication, please cite*:
Luebbert, L., & Pachter, L. (2023). Efficient querying of genomic reference databases with gget. Bioinformatics. https://doi.org/10.1093/bioinformatics/btac836
Read the article here: https://doi.org/10.1093/bioinformatics/btac836
Installation
bash
uv pip install gget
or
bash
pip install --upgrade gget
For use in Jupyter Lab / Google Colab: ```python
Python
import gget ```
🔗 Manual
🪄 Quick start guide
Command line: ```bash
Fetch all Homo sapiens reference and annotation FTPs from the latest Ensembl release
$ gget ref homo_sapiens
Get Ensembl IDs of human genes with "ace2" or "angiotensin converting enzyme 2" in their name/description
$ gget search -s homo_sapiens 'ace2' 'angiotensin converting enzyme 2'
Look up gene ENSG00000130234 (ACE2) and its transcript ENST00000252519
$ gget info ENSG00000130234 ENST00000252519
Fetch the amino acid sequence of the canonical transcript of gene ENSG00000130234
$ gget seq --translate ENSG00000130234
Quickly find the genomic location of (the start of) that amino acid sequence
$ gget blat MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS
BLAST (the start of) that amino acid sequence
$ gget blast MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS
Align multiple nucleotide or amino acid sequences against each other (also accepts path to FASTA file)
$ gget muscle MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS MSSSSWLLLSLVEVTAAQSTIEQQAKTFLDKFHEAEDLFYQSLLAS
Align one or more amino acid sequences against a reference (containing one or more sequences) (local BLAST) (also accepts paths to FASTA files)
$ gget diamond MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS -ref MSSSSWLLLSLVEVTAAQSTIEQQAKTFLDKFHEAEDLFYQSLLAS
Use Enrichr for an ontology analysis of a list of genes
$ gget enrichr -db ontology ACE2 AGT AGTR1 ACE AGTRAP AGTR2 ACE3P
Get the human tissue expression of gene ACE2
$ gget archs4 -w tissue ACE2
Get the protein structure (in PDB format) of ACE2 as stored in the Protein Data Bank (PDB ID returned by gget info)
$ gget pdb 1R42 -o 1R42.pdb
Find Eukaryotic Linear Motifs (ELMs) in a protein sequence
$ gget setup elm # setup only needs to be run once $ gget elm -o results MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS
Fetch a scRNAseq count matrix (AnnData format) based on specified gene(s), tissue(s), and cell type(s) (default species: human)
$ gget setup cellxgene # setup only needs to be run once $ gget cellxgene --gene ACE2 SLC5A1 --tissue lung --celltype 'mucus secreting cell' -o exampleadata.h5ad
Predict the protein structure of GFP from its amino acid sequence
$ gget setup alphafold # setup only needs to be run once
$ gget alphafold MSKGEELFTGVVPILVELDGDVNGHKFSVSGEGEGDATYGKLTLKFICTTGKLPVPWPTLVTTFSYGVQCFSRYPDHMKQHDFFKSAMPEGYVQERTIFFKDDGNYKTRAEVKFEGDTLVNRIELKGIDFKEDGNILGHKLEYNYNSHNVYIMADKQKNGIKVNFKIRHNIEDGSVQLADHYQQNTPIGDGPVLLPDNHYLSTQSALSKDPNEKRDHMVLLEFVTAAGITHGMDELYK
Python (Jupyter Lab / Google Colab):
python
import gget
gget.ref("homosapiens")
gget.search(["ace2", "angiotensin converting enzyme 2"], "homosapiens")
gget.info(["ENSG00000130234", "ENST00000252519"])
gget.seq("ENSG00000130234", translate=True)
gget.blat("MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS")
gget.blast("MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS")
gget.muscle(["MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS", "MSSSSWLLLSLVEVTAAQSTIEQQAKTFLDKFHEAEDLFYQSLLAS"])
gget.diamond("MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS", reference="MSSSSWLLLSLVEVTAAQSTIEQQAKTFLDKFHEAEDLFYQSLLAS")
gget.enrichr(["ACE2", "AGT", "AGTR1", "ACE", "AGTRAP", "AGTR2", "ACE3P"], database="ontology", plot=True)
gget.archs4("ACE2", which="tissue")
gget.pdb("1R42", save=True)
gget.setup("elm") # setup only needs to be run once orthodf, regexdf = gget.elm("MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS")
gget.setup("cellxgene") # setup only needs to be run once gget.cellxgene(gene = ["ACE2", "SLC5A1"], tissue = "lung", cell_type = "mucus secreting cell")
gget.setup("alphafold") # setup only needs to be run once
gget.alphafold("MSKGEELFTGVVPILVELDGDVNGHKFSVSGEGEGDATYGKLTLKFICTTGKLPVPWPTLVTTFSYGVQCFSRYPDHMKQHDFFKSAMPEGYVQERTIFFKDDGNYKTRAEVKFEGDTLVNRIELKGIDFKEDGNILGHKLEYNYNSHNVYIMADKQKNGIKVNFKIRHNIEDGSVQLADHYQQNTPIGDGPVLLPDNHYLSTQSALSKDPNEKRDHMVLLEFVTAAGITHGMDELYK")
Call `gget` from R using [reticulate](https://rstudio.github.io/reticulate/):
r
system("pip install gget")
install.packages("reticulate")
library(reticulate)
gget <- import("gget")
gget$ref("homosapiens") gget$search(list("ace2", "angiotensin converting enzyme 2"), "homosapiens") gget$info(list("ENSG00000130234", "ENST00000252519")) gget$seq("ENSG00000130234", translate=TRUE) gget$blat("MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS") gget$blast("MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS") gget$muscle(list("MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS", "MSSSSWLLLSLVEVTAAQSTIEQQAKTFLDKFHEAEDLFYQSLLAS"), out="out.afa") gget$diamond("MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS", reference="MSSSSWLLLSLVEVTAAQSTIEQQAKTFLDKFHEAEDLFYQSLLAS") gget$enrichr(list("ACE2", "AGT", "AGTR1", "ACE", "AGTRAP", "AGTR2", "ACE3P"), database="ontology") gget$archs4("ACE2", which="tissue") gget$pdb("1R42", save=TRUE) ```
More tutorials
Owner
- Name: Pachter Lab
- Login: pachterlab
- Kind: organization
- Email: lpachter@caltech.edu
- Location: Pasadena, CA
- Website: http://pachterlab.github.io
- Repositories: 128
- Profile: https://github.com/pachterlab
Citation (CITATION.cff)
cff-version: 1.2.0
message: "If you use gget in a publication, please cite it as below."
authors:
- family-names: "Luebbert"
given-names: "Laura"
orcid: "https://orcid.org/0000-0003-1379-2927"
- family-names: "Pachter"
given-names: "Lior"
orcid: "https://orcid.org/0000-0002-9164-6231"
title: "gget"
# version: 0.2.6
doi: "10.1093/bioinformatics/btac836"
# date-released: 2022
url: "https://github.com/pachterlab/gget"
preferred-citation:
type: article
authors:
- family-names: "Luebbert"
given-names: "Laura"
orcid: "https://orcid.org/0000-0003-1379-2927"
- family-names: "Pachter"
given-names: "Lior"
orcid: "https://orcid.org/0000-0002-9164-6231"
doi: "10.1093/bioinformatics/btac836"
journal: "Bioinformatics"
# month: 9
# start: 1 # First page number
# end: 10 # Last page number
title: "Efficient querying of genomic reference databases with gget"
# issue: 1
# volume: 1
year: 2023
GitHub Events
Total
- Create event: 4
- Release event: 2
- Issues event: 15
- Watch event: 113
- Delete event: 1
- Member event: 2
- Issue comment event: 25
- Push event: 297
- Pull request event: 13
- Fork event: 7
Last Year
- Create event: 4
- Release event: 2
- Issues event: 15
- Watch event: 113
- Delete event: 1
- Member event: 2
- Issue comment event: 25
- Push event: 297
- Pull request event: 13
- Fork event: 7
Committers
Last synced: 9 months ago
Top Committers
| Name | Commits | |
|---|---|---|
| Laura Luebbert | 5****t | 2,213 |
| choang | c****g@c****u | 397 |
| techno-sam | 7****m | 82 |
| josephrich98 | j****8@g****m | 72 |
| Chi Hoang | 3****2 | 12 |
| Victor Garcia Ruiz | 5****5 | 9 |
| vecerkovakaterina | v****1@g****m | 6 |
| Gavin John | g****n@g****m | 4 |
| Lior Pachter | l****r@g****m | 3 |
| noriakis | 3****s | 3 |
| DylanLawless | d****s@e****h | 2 |
| Arman | 3****n | 1 |
| Austin | a****1@g****m | 1 |
| Christian Brueffer | c****n@b****o | 1 |
| JJ | 1****s | 1 |
| Nils Homer | n****3 | 1 |
| Tomás Di Domenico | t****o@t****u | 1 |
| Mayuko Boffelli | m****i@u****u | 1 |
Committer Domains (Top 20 + Academic)
Issues and Pull Requests
Last synced: 6 months ago
All Time
- Total issues: 75
- Total pull requests: 99
- Average time to close issues: 2 months
- Average time to close pull requests: 8 days
- Total issue authors: 52
- Total pull request authors: 17
- Average comments per issue: 2.53
- Average comments per pull request: 0.32
- Merged pull requests: 93
- Bot issues: 0
- Bot pull requests: 0
Past Year
- Issues: 12
- Pull requests: 16
- Average time to close issues: about 2 months
- Average time to close pull requests: 21 days
- Issue authors: 7
- Pull request authors: 6
- Average comments per issue: 1.58
- Average comments per pull request: 0.63
- Merged pull requests: 13
- Bot issues: 0
- Bot pull requests: 0
Top Authors
Issue Authors
- lauraluebbert (16)
- lakigigar (4)
- abearab (3)
- KleinSamuel (2)
- gouinK (2)
- adeslatt (2)
- almart7 (2)
- JackCurragh (1)
- Paulie-ai (1)
- vkullu (1)
- alexpreynolds (1)
- nahid18 (1)
- ahwchemistry (1)
- atolopko-czi (1)
- nick-youngblut (1)
Pull Request Authors
- lauraluebbert (64)
- techno-sam (23)
- anhchi172 (12)
- austinv11 (4)
- Pandapip1 (4)
- josephrich98 (3)
- abearab (3)
- jkhales (2)
- mboffelli (2)
- vecerkovakaterina (2)
- AubakirovArman (2)
- victorg775 (1)
- cbrueffer (1)
- nh13 (1)
- noriakis (1)
Top Labels
Issue Labels
Pull Request Labels
Packages
- Total packages: 2
-
Total downloads:
- pypi 30,656 last-month
- Total docker downloads: 58
-
Total dependent packages: 4
(may contain duplicates) -
Total dependent repositories: 3
(may contain duplicates) - Total versions: 100
- Total maintainers: 1
pypi.org: gget
Efficient querying of genomic databases.
- Homepage: https://github.com/pachterlab/gget
- Documentation: https://gget.readthedocs.io/
- License: BSD-2
-
Latest release: 0.29.2
published 8 months ago
Rankings
Maintainers (1)
proxy.golang.org: github.com/pachterlab/gget
- Documentation: https://pkg.go.dev/github.com/pachterlab/gget#section-documentation
- License: bsd-2-clause
-
Latest release: v0.29.2
published 8 months ago
Rankings
Dependencies
- actions/checkout main composite
- actions/setup-python v1 composite
- actions/checkout v2 composite
- EndBug/add-and-commit v4 composite
- actions/checkout v2 composite
- sangonzal/repository-traffic-action v.0.1.6 composite
- coverage >=5.1 development
- lxml * development
- pytest >=7.0.0 development
- beautifulsoup4 >=4.10.0
- ipython *
- ipywidgets *
- matplotlib *
- mysql-connector-python >=8.0.5,<=8.0.29
- numpy >=1.17.2
- pandas >=1.0.0
- py3Dmol >=1.8.0
- requests >=2.22.0
- tqdm *




