alphastats
Python Package for the downstream analysis of mass-spectrometry-based proteomics data
Science Score: 39.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
○CITATION.cff file
-
✓codemeta.json file
Found codemeta.json file -
✓.zenodo.json file
Found .zenodo.json file -
✓DOI references
Found 4 DOI reference(s) in README -
○Academic publication links
-
○Committers with academic emails
-
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (15.6%) to scientific vocabulary
Keywords
Keywords from Contributors
Repository
Python Package for the downstream analysis of mass-spectrometry-based proteomics data
Basic Info
- Host: GitHub
- Owner: MannLabs
- License: apache-2.0
- Language: Jupyter Notebook
- Default Branch: main
- Homepage: https://alphapeptstats.readthedocs.io/en/latest/
- Size: 118 MB
Statistics
- Stars: 73
- Watchers: 5
- Forks: 16
- Open Issues: 17
- Releases: 33
Topics
Metadata Files
README.md
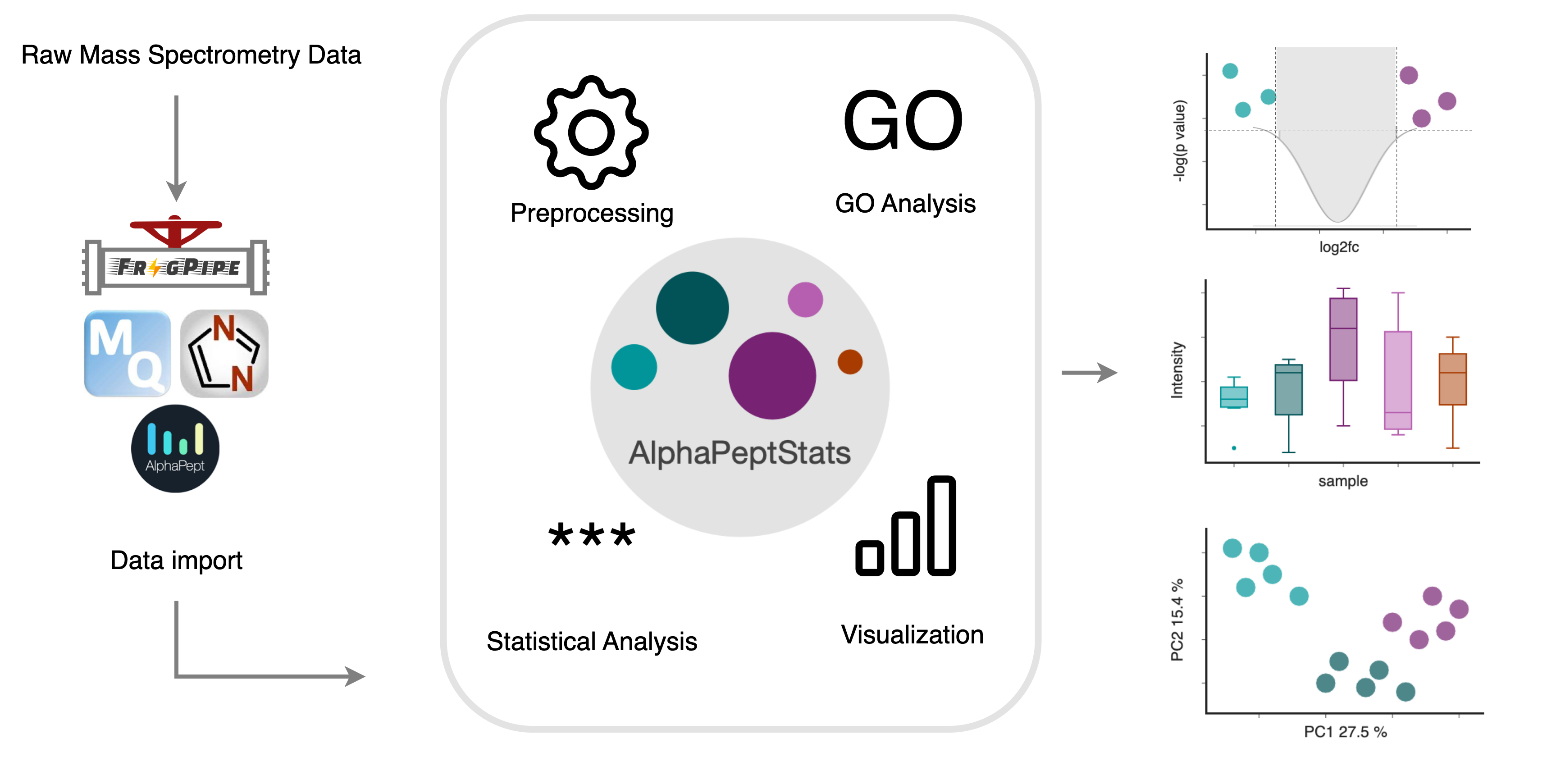
An open-source Python package for mass spectrometry downstream data analysis from the Mann Group at the University of Copenhagen and the Mann Group at the MPI Biochemistry.
Check out alphapept.org for other packages of AlphaPept ecosystem.
GUI Preview

=> Run the app right now in your browser
Installation
AlphaPeptStats can be used * via a Graphical User Interface, * as a python library, or * as a Docker container.
One Click Installer
One-click installers for MacOS, Windows and Linux can be found here.
Windows
Download the latest alphastats-X.Y.Z-windows-amd64.exe build and double click it to install. If you receive a warning during installation click Run anyway.
Important note: always install AlphaPeptStats into a new folder, as the installer will not properly overwrite existing installations.
Linux
Download the latest alphastats-X.Y.Z-linux-x64.deb build and install it via dpkg -i alphastats-X.Y.Z-linux-x64.deb.
MacOS
Download the latest build suitable for your chip architecture
(can be looked up by clicking on the Apple Symbol > About this Mac > Chip ("M1", "M2", "M3" -> arm64, "Intel" -> x64),
alphastats-X.Y.Z-macos-darwin-arm64.pkg or alphastats-X.Y.Z-macos-darwin-x64.pkg. Open the parent folder of the downloaded file in Finder,
right-click and select open. If you receive a warning during installation click Open.
In newer MacOS versions, additional steps are required to enable installation of unverified software.
This is indicated by a dialog telling you alphastats. ... .pkg Not Opened.
1. Close this dialog by clicking Done.
2. Choose Apple menu > System Settings, then Privacy & Security in the sidebar. (You may need to scroll down.)
3. Go to Security, locate the line "alphadia.pkg was blocked to protect your Mac" then click Open Anyway.
4. In the dialog windows, click Open Anyway.
Pip Installation
AlphaStats can be installed in an existing Python >=3.9 environment with a single bash command.
bash
pip install alphastats
In case you want to use the Graphical User Interface, use following command in the command line:
bash
alphastats gui
If you get an AxiosError: Request failed with status code 403' when uploading files, try running DISABLE_XSRF=1 alphastats gui.
If you receive an error like library 'hdf5' not found, your computer is missing the HDF5 library. Install it via your favorite package manager or use conda create --name alphastats python=3.9 hdf5.
Alternatively, use conda install -c anaconda pytables.
AlphaStats can be imported as a Python package into any Python script or notebook with the command import alphastats.
A brief Jupyter notebook tutorial on how to use the API is also present in the nbs folder.
Docker version
The containerized version can be used to run alphapeptstats without any installation (apart from Docker)
1. Setting up Docker
Install the latest version of docker (https://docs.docker.com/engine/install/).
2. Start the container
bash
PORT=8501
SESSIONS_PATH=./sessions
docker run -p $PORT:8501 -v $SESSIONS_PATH:/app/sessions mannlabs/alphastats:latest
After initial download of the container, alphapeptstats will start running on http://localhost:$PORT.
Note: this will create a directory $SESSIONS_PATH where sessions will be stored.
API Documentation
AlphaPeptStats provides an extensive API documentation.
Troubleshooting
In case of issues, check out the following:
- Issues: Try a few different search terms to find out if a similar problem has been encountered before
Common problems
How to resolve " error: Microsoft Visual C++ 14.0 or greater is required. Get it with "Microsoft C++ Build Tools" " ?
Please, find a description on how to update required tools here.
How to resolve "ERROR: Could not find a local HDF5 installation" on Mac Silicon (M1/M2/M3)?
Before installing AlphaPeptStats you might need to install pytables first:
conda install -c anaconda pytables
License
AlphaStats was developed by the Mann Group at the University of Copenhagen and is now maintained by the Mann Group at the MPI Biochemistry. It is freely available with an Apache License. External Python packages have their own licenses, which can be consulted on their respective websites.
How to contribute
If you like this software, you can give us a star to boost our visibility! All direct contributions are also welcome. Feel free to post a new issue or clone the repository and create a pull request with a new branch. For an even more interactive participation, check out the discussions and the Contributors License Agreement.
Notes for developers
Tagging of changes
In order to have release notes automatically generated, changes need to be tagged with labels.
The following labels are used (should be safe-explanatory):
breaking-change, bug, enhancement.
Release a new version
This package uses a shared release process defined in the alphashared repository. Please see the instructions there.
pre-commit hooks
It is highly recommended to use the provided pre-commit hooks, as the CI pipeline enforces all checks therein to pass in order to merge a branch.
The hooks need to be installed once by
bash
pip install -r requirements_dev.txt
pre-commit install
You can run the checks yourself using:
bash
pre-commit run --all-files
The detect-secrets hook fails
This is because you added some code that was identified as a potential secret.
1. Run detect-secrets scan --exclude-files testfiles --exclude-lines '"(hash|id|image/\w+)":.*' > .secrets.baseline
(check .pre-commit-config.yaml for the exact parameters)
2. Run detect-secrets audit .secrets.baseline and check if the detected 'secret' is actually a secret
3. Commit the latest version of .secrets.baseline
Changelog
See the GitHub Release Notes for changes from version 0.6.8 on, HISTORY.md for older versions.
Citation
Citation:
Krismer, E., Bludau, I., Strauss M. & Mann M. (2023). AlphaPeptStats: an open-source Python package for automated and scalable statistical analysis of mass spectrometry-based proteomics. Bioinformatics https://doi.org/10.1093/bioinformatics/btad461
Owner
- Name: Mann Labs
- Login: MannLabs
- Kind: organization
- Repositories: 16
- Profile: https://github.com/MannLabs
GitHub Events
Total
- Create event: 90
- Release event: 1
- Issues event: 24
- Watch event: 16
- Delete event: 96
- Member event: 2
- Issue comment event: 71
- Push event: 450
- Pull request review comment event: 660
- Pull request review event: 586
- Pull request event: 184
- Fork event: 2
Last Year
- Create event: 90
- Release event: 1
- Issues event: 24
- Watch event: 16
- Delete event: 96
- Member event: 2
- Issue comment event: 71
- Push event: 450
- Pull request review comment event: 660
- Pull request review event: 586
- Pull request event: 184
- Fork event: 2
Committers
Last synced: over 1 year ago
Top Committers
| Name | Commits | |
|---|---|---|
| elena-krismer | e****r@h****m | 582 |
| Elena Krismer | 7****r | 40 |
| Mikhail Lebedev | l****l@o****m | 10 |
| Mikhail Lebedev | 4****t | 3 |
| dependabot[bot] | 4****] | 2 |
| Maximilian Strauss | s****n@g****m | 1 |
| ibludau | i****u@g****m | 1 |
Issues and Pull Requests
Last synced: 6 months ago
All Time
- Total issues: 41
- Total pull requests: 392
- Average time to close issues: 8 months
- Average time to close pull requests: 23 days
- Total issue authors: 24
- Total pull request authors: 8
- Average comments per issue: 1.07
- Average comments per pull request: 0.77
- Merged pull requests: 250
- Bot issues: 0
- Bot pull requests: 133
Past Year
- Issues: 14
- Pull requests: 215
- Average time to close issues: 5 months
- Average time to close pull requests: 25 days
- Issue authors: 10
- Pull request authors: 6
- Average comments per issue: 0.5
- Average comments per pull request: 0.42
- Merged pull requests: 181
- Bot issues: 0
- Bot pull requests: 18
Top Authors
Issue Authors
- elena-krismer (4)
- JohnSuberu (3)
- acesnik (3)
- steph-robinson (3)
- JM-Bader (3)
- mschwoer (2)
- straussmaximilian (2)
- KunHHE (2)
- andzajan (2)
- glycoaddict (1)
- bolak92 (1)
- amptsmb (1)
- michaelsteidel86 (1)
- JuliaS92 (1)
- pejota66 (1)
Pull Request Authors
- mschwoer (179)
- dependabot[bot] (132)
- JuliaS92 (97)
- elena-krismer (45)
- boopthesnoot (22)
- github-actions[bot] (5)
- PatriciaSkowronek (4)
- ibludau (2)
Top Labels
Issue Labels
Pull Request Labels
Packages
- Total packages: 1
-
Total downloads:
- pypi 360 last-month
- Total dependent packages: 0
- Total dependent repositories: 1
- Total versions: 37
- Total maintainers: 1
pypi.org: alphastats
An open-source Python package for automated and scalable statistical analysis of mass spectrometry-based proteomics
- Homepage: https://github.com/MannLabs/alphastats
- Documentation: https://mannlabs.github.io/alphapeptstats/
- License: Apache
-
Latest release: 0.7.1
published 7 months ago
Rankings
Maintainers (1)
Dependencies
- actions/checkout v2 composite
- conda-incubator/setup-miniconda v2 composite
- pypa/gh-action-pypi-publish master composite
- actions/checkout v3 composite
- actions/setup-python v3 composite
- codecov/codecov-action v2 composite
- actions/checkout v2 composite
- actions/create-release v1 composite
- actions/upload-release-asset v1 composite
- conda-incubator/setup-miniconda v2 composite
- python 3.8-slim build
- alphastats ==0.6.3
- altair ==4.2.0
- anndata ==0.8.0
- attrs ==22.1.0
- batchglm ==0.7.4
- beautifulsoup4 ==4.11.1
- bleach ==5.0.1
- blinker ==1.5
- cachetools ==5.2.0
- click ==8.0.1
- cloudpickle ==2.2.0
- commonmark ==0.9.1
- contourpy ==1.0.5
- cycler ==0.11.0
- dask ==2022.10.0
- data_cache ==0.1.6
- decorator ==5.1.1
- defusedxml ==0.7.1
- diffxpy ==0.7.4
- entrypoints ==0.4
- et-xmlfile ==1.1.0
- fastjsonschema ==2.16.2
- fonttools ==4.38.0
- fsspec ==2022.10.0
- gitdb ==4.0.9
- gitpython ==3.1.32
- h5py ==3.7.0
- importlib-metadata ==5.0.0
- iteration_utilities ==0.11.0
- joblib ==1.2.0
- jsonschema ==4.16.0
- jupyter-client ==7.4.3
- jupyter-core ==4.11.2
- jupyterlab-pygments ==0.2.2
- kiwisolver ==1.4.4
- lazy-loader ==0.1rc2
- littleutils ==0.2.2
- llvmlite ==0.39.1
- locket ==1.0.0
- markdown-it-py ==2.1.0
- matplotlib ==3.6.0
- mdit-py-plugins ==0.3.1
- mdurl ==0.1.2
- mistune ==2.0.4
- myst_parser ==0.18.1
- natsort ==8.2.0
- nbclient ==0.7.0
- nbconvert ==7.2.2
- nbformat ==5.7.0
- nbsphinx ==0.8.9
- nest-asyncio ==1.5.6
- numba ==0.56.4
- numexpr ==2.8.3
- numpy ==1.23.5
- openpyxl ==3.0.10
- outdated ==0.2.1
- pandas ==2.0.0
- pandas_flavor ==0.3.0
- pandoc ==2.2
- pandocfilters ==1.5.0
- partd ==1.3.0
- patsy ==0.5.3
- pillow ==9.2.0
- pingouin ==0.5.3
- plotly ==5.11.0
- plumbum ==1.8.0
- ply ==3.11
- protobuf ==3.20.3
- pyarrow ==9.0.0
- pydeck ==0.8.0b4
- pympler ==1.0.1
- pynndescent ==0.5.7
- pyrsistent ==0.18.1
- python-dateutil ==2.8.2
- pyyaml ==6.0
- pyzmq ==24.0.1
- rich ==12.6.0
- scikit-learn ==1.2.1
- scipy ==1.10.1
- seaborn ==0.12.1
- semver ==2.13.0
- six ==1.16.0
- sklearn ==0.0
- sklearn_pandas ==2.2.0
- smmap ==5.0.0
- sparse ==0.13.0
- sphinx-argparse ==0.3.2
- sphinx-autodoc-typehints ==1.19.4
- sphinx-rtd-theme ==1.0.0
- statsmodels ==0.13.5
- streamlit ==1.22.0
- tables ==3.7.0
- tabulate ==0.9.0
- tenacity ==8.1.0
- threadpoolctl ==3.1.0
- tinycss2 ==1.2.1
- toml ==0.10.2
- toolz ==0.12.0
- tornado ==6.2
- tqdm ==4.64.1
- traitlets ==5.5.0
- typing-extensions ==4.4.0
- tzdata ==2022.5
- tzlocal ==4.2
- umap-learn ==0.5.3
- validators ==0.20.0
- watchdog ==2.1.9
- webencodings ==0.5.1
- xarray ==2022.10.0
- zipp ==3.10.0
- anndata ==0.9.1
- click ==8.0.1
- combat ==0.3.3
- data_cache >=0.1.6
- diffxpy ==0.7.4
- kaleido ==0.2.1
- nbformat >=5.0
- numba ==0.56.4
- numba-stats ==0.5.0
- numpy ==1.23.5
- openpyxl >=3.0.10
- pandas ==2.0.2
- pingouin ==0.5.3
- plotly ==5.15.0
- pyteomics ==4.6.0
- scikit-learn ==1.2.2
- scipy ==1.10.1
- sklearn_pandas ==2.2.0
- statsmodels ==0.14.0
- streamlit ==1.22.0
- swifter ==1.2.0
- tables ==3.7.0
- tqdm >=4.64.0
- umap-learn ==0.5.3
- xlsxwriter ==3.1.0

