Science Score: 67.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
✓CITATION.cff file
Found CITATION.cff file -
✓codemeta.json file
Found codemeta.json file -
✓.zenodo.json file
Found .zenodo.json file -
✓DOI references
Found 2 DOI reference(s) in README -
○Academic publication links
-
✓Committers with academic emails
1 of 2 committers (50.0%) from academic institutions -
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (14.4%) to scientific vocabulary
Keywords
Repository
Multi-view clustering with flexible ensemble structures.
Basic Info
- Host: GitHub
- Owner: mdbloice
- License: mit
- Language: Python
- Default Branch: master
- Homepage: https://pyrea.readthedocs.io
- Size: 230 KB
Statistics
- Stars: 24
- Watchers: 2
- Forks: 3
- Open Issues: 1
- Releases: 0
Topics
Metadata Files
README.md
Pyrea

Multi-view clustering with flexible ensemble structures.
The name Pyrea is derived from the Greek word Parea, meaning a group of friends who gather to share experiences, values, and ideas.
Installation
Install Pyrea using pip:
bash
pip install pyrea
This will install the latest version of Pyrea from PyPI.
Demonstration Notebooks
A demonstration of Pyrea's usage on the Nutrimouse dataset can be found in the following Jupyter notebook:
In this notebook, hierarchical and spectral clustering are performed on the Nutrimouse multi-view dataset, tuned using Pyrea's genetic algorithm functionality.
More notebooks will be added in due course.
Usage
The Pyrea software package is the accompanying software for our paper:
Pfeifer, B., Bloice, M.D., & Schimek, M.G. (2023). Parea: multi-view ensemble clustering for cancer subtype discovery. Journal of Biomedical Informatics. https://doi.org/10.1016/j.jbi.2023.104406
While Pyrea allows for flexible and custom architectures to be built, two structures are discussed specifically in the paper cited above, namely Parea 1 and Parea 2.
Both the structures, which are described in detail below as well as in the paper mentioned above, can be quickly generated and applied to your data using two helper functions, parea_1() and parea_2(), and can be quickly run as follows:
```python import pyrea import numpy as np
Create sample data:
d1 = np.random.rand(100,10) d2 = np.random.rand(100,10) d3 = np.random.rand(100,10)
data = [d1,d2, d3]
labels = pyrea.parea_2(data) ```
which executes Parea 2.
Default parameters are used which match those used in our experiments discussed in the paper referenced above. These default parameters can of course be overridden. As there are many combinations of parameters that could be used, a genetic algorithm can be utilised to find the optimum parameters, as shown in the next section.
Genetic Algorithm
The Parea 1 and Parea 2 structures can be optimised using a genetic algorithm in order to find the best combinations of clustering methods, fusion methods, and number of clusters.
For example, to find optimal parameters for Parea 2:
```python import pyrea from sklearn import datasets
d1 = datasets.loadiris().data d2 = datasets.loadiris().data d3 = datasets.load_iris().data
data = [d1,d2,d3]
params = pyrea.parea2genetic(data, kmin=2, kmax=5) ```
where k_min and k_max refer to the minimum and maximum number of clusters to attempt for each layer, respectively.
Note that params contains the optimal parameters found by the genetic algorithm. To get the labels, run parea_2() passing your data and these optimal parameters:
python
pyrea.parea_2(data, *params)
which will return the cluster labels for your data.
Also, you may choose to define the final number of clusters returned by the algorithm (but allowing it to optimise intermediate numbers of clusters) by defining k_final, e.g:
python
params = pyrea.parea_2_genetic(data, k_min=2, k_max=5, k_final=3)
and calling pyrea_2() as follows:
python
pyrea.parea_2(data, params, k_final=3)
Genetic Algorithm Update
The genetic algorithm functions now support arbitrary numbers of views, and the population and number of generations can now be adjusted. See this notebook for a demonstration of this usage on the Nutrimouse dataset.
API
Please note that Pyrea is work in progress. The API may change from version to version and introduce breaking changes.
In Pyrea, your data are organised in to views. A view consists of the data in the form of a 2D matrix, and an associated clustering algorithm (a clusterer).
To create a view you must have some data, and a clusterer:
```python import pyrea
Create your data, which must be a 2-dimensional array/matrix.
d = [[1,2,3], [4,5,6], [7,8,9]]
Create a clusterer
c = pyrea.clusterer("hierarchical", n_clusters=2, method='ward')
v = pyrea.view(d, c) ```
You now have a view v, containing the data d using the clustering algorithm
c. Note that many views can share the same clusterer, or each view may have a
unique clusterer.
To obtain the cluster solution the specified view can be executed
python
v.execute()
The clustering algorithm can be either 'spectral', 'hierarchical', 'dbscan', or 'optics'. See the documentation for a complete list of parameters that can be passed when creating a clusterer.
As this is a library for multi-view ensemble learning, you will normally have multiple views.
A fusion algorithm is therefore used to fuse the clusterings created from multiple views. Therefore, our next step is to create a fuser object:
python
f = pyrea.fuser('disagreement')
With you fusion algorithm f, you can execute an ensemble. The ensemble is created with a set of views and a fusion algorithm,
and its returned object (distance or affinity matrix) can again be specified as a view:
```python
Create a new clusterer with precomputed=True
cpre = pyrea.clusterer("hierarchical", nclusters=2, method='ward', precomputed=True) vres = pyrea.view(pyrea.executeensemble([v1, v2, v3], f), c_pre) ```
This newly created view, v_res can subsequently be fed into another ensemble,
allowing you to create stacked ensemble architectures, with high flexibility.
A full example is shown below, using random data:
```python import pyrea import numpy as np
Create two datasets with random values of 1000 samples and 100 features per sample.
d1 = np.random.rand(1000,100) d2 = np.random.rand(1000,100)
Define the clustering algorithm(s) you want to use. In this case we used the same
algorithm for both views. By default n_clusters=2.
c = pyrea.clusterer('hierarchical', n_clusters=2, method='ward')
Create the views using the data and the same clusterer
v1 = pyrea.view(d1, c) v2 = pyrea.view(d2, c)
Create a fusion object
f = pyrea.fuser('disagreement')
Specify a clustering algorithm (precomputed = True)
cpre = pyrea.clusterer("hierarchical", nclusters=2, method='ward', precomputed=True)
Execute an ensemble based on your views and a fusion algorithm
vres = pyrea.view(pyrea.executeensemble([v1, v2], f), c_pre)
The cluster solution can be obtained as follows
v_res.execute() ```
Ensemble Structures
Complex structures can be built using Pyrea.
For example, examine the two structures below:
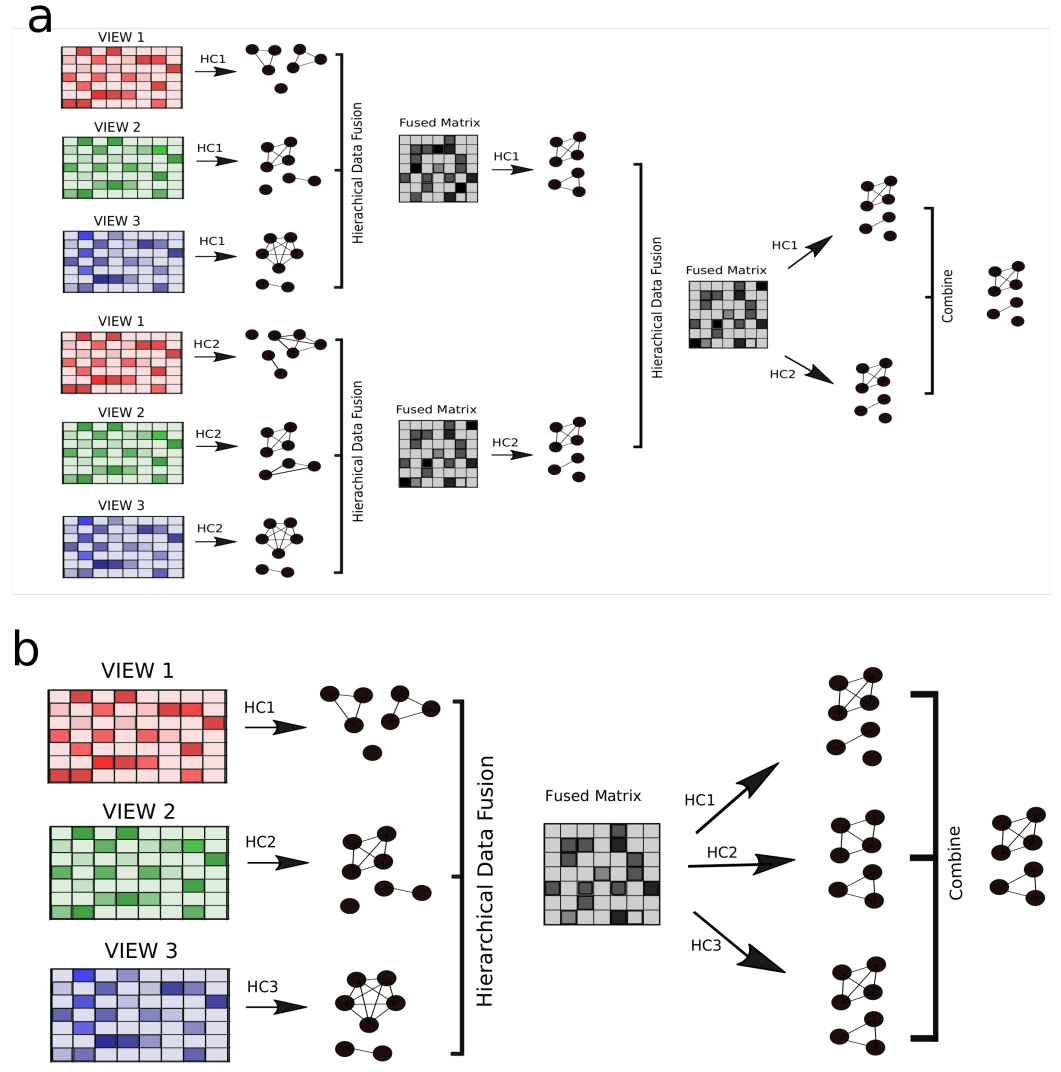
We will demonstrate how to create deep and flexible ensemble structures using the examples a) and b) from the image above.
Example A
This ensemble consists of two sets of three views, which are clustered, fused, and then once again combined in a second layer.
We create two ensembles, which represent the first layer of structure a) in the image above:
```python import pyrea import numpy as np
Clusterers:
hc1 = pyrea.clusterer('hierarchical', method='ward', nclusters=2) hc2 = pyrea.clusterer('hierarchical', method='complete', nclusters=2)
Fusion algorithm:
f = pyrea.fuser('disagreement')
Create three random datasets
d1 = np.random.rand(100,10) d2 = np.random.rand(100,10) d3 = np.random.rand(100,10)
Views for ensemble 1
v1 = pyrea.view(d1, hc1) v2 = pyrea.view(d2, hc1) v3 = pyrea.view(d3, hc1)
Execute ensemble 1 and retrieve a new view, which is used later.
hc1pre = pyrea.clusterer('hierarchical', method='ward', nclusters=2, precomputed=True) vensemble1 = pyrea.view(pyrea.executeensemble([v1, v2, v3], f), hc1pre)
Views for ensemble 2
v4 = pyrea.view(d1, hc2) v5 = pyrea.view(d2, hc2) v6 = pyrea.view(d3, hc2)
Execute our second ensemble, and retreive a new view:
hc2pre = pyrea.clusterer('hierarchical', method='complete', nclusters=2, precomputed=True) vensemble2 = pyrea.view(pyrea.executeensemble([v4, v5, v6], f), hc2pre)
Now we can execute a further ensemble, using the views generated from the
two previous ensemble methods:
dfuse = pyrea.executeensemble([vensemble1, vensemble2], f)
The returned distance matrix is now used as an input for the two clustering methods (hc1 and hc2)
v1fuse = pyrea.view(dfuse, hc1pre) v2fuse = pyrea.view(dfuse, hc2pre)
and the cluster solutions are combined
pyrea.consensus([v1fuse.execute(), v2fuse.execute()]) ```
Helper Function
See the parea_1() helper function for a pre-built version of structure above.
Example B
As for structure b) in the image above, this can implemented as follows:
```python import pyrea import numpy as np
Clustering algorithms
c1 = pyrea.clusterer('hierarchical', method='ward', nclusters=2) c2 = pyrea.clusterer('hierarchical', method='complete', nclusters=2) c3 = pyrea.clusterer('hierarchical', method='single', n_clusters=2)
Clustering algorithms (so it works with a precomputed distance matrix)
c1pre = pyrea.clusterer('hierarchical', method='ward', nclusters=2, precomputed=True) c2pre = pyrea.clusterer('hierarchical', method='complete', nclusters=2, precomputed=True) c3pre = pyrea.clusterer('hierarchical', method='single', nclusters=2, precomputed=True)
Fusion algorithm
f = pyrea.fuser('disagreement')
Create the views with the random data directly:
v1 = pyrea.view(np.random.rand(100,10), c1) v2 = pyrea.view(np.random.rand(100,10), c2) v3 = pyrea.view(np.random.rand(100,10), c3)
Create the ensemble and define new views based on the returned disagreement matrix v_res
vres = pyrea.executeensemble([v1, v2, v3], f) v1res = pyrea.view(vres, c1pre) v2res = pyrea.view(vres, c2pre) v3res = pyrea.view(vres, c3_pre)
Get the final cluster solution
pyrea.consensus([v1res.execute(), v2res.execute(), v3_res.execute()]) ```
Helper Function
See the parea_2() helper function for a pre-built version of structure above.
Extensible
Pyrea has been designed to be extensible. It allows you to use Pyrea's data fusion techniques with custom clustering algorithms that can be loaded in to Pyrea at run-time.
By providing a View with a ClusterMethod object, it makes providing custom clustering algorithms uncomplicated. See Extending Pyrea for details.
Work In Progress and Future Work
Several features are currently work in progress, future updates will include the features described in the sections below.
HCfused Clustering Algorithm
A novel fusion technique, developed by one of the authors of this software package, named HCfused, will be included soon in a future update.
General Genetic Optimisation
The package will be extended to allow for any custom Pyrea structures to be optimised using a genetic algorithm.
Compilation of HC Fused C++ Code
To use the HC Fused method you may need to compile the source code yourself if binaries are not available for your operating system. HC Fused has been implemented in C++, see the HC_fused_cpp_opt6.cpp source file for more details.
Pre-compiled binaries are available for Linux and have been tested using Linux only. The instructions below pertain to Linux only. For Windows please consult https://docs.python.org/3.5/library/ctypes.html#loading-shared-libraries and use a compiler such as MSVC or MinGW.
To compile HC Fused (and then create a shared library/dynamic library) execute the following on the command line:
bash
$ clang++ -c -fPIC HC_fused_cpp_opt6.cpp -o HC_fused_cpp_opt6.o
and then create the .so file shared library file:
bash
$ clang++ HC_fused_cpp_opt6.o -shared -o libhcfused.so
and finally place the libhcfused.so file in the root directory of the package's installation directory.
Tests
Installation is tested using Python versions 3.8, 3.9, 3.10, and 3.11 on Ubuntu 20.04 LTS only. See the project's Actions for details. The package should also work using Python 3.6 and 3.7 on other operating systems, however.
Miscellaneous
Logo made by Adobe Express Logo Maker: https://www.adobe.com/express/create/logo
Owner
- Name: Marcus D. Bloice
- Login: mdbloice
- Kind: user
- Location: Graz, Austria
- Company: Medical University Graz
- Website: https://imi.medunigraz.at
- Repositories: 3
- Profile: https://github.com/mdbloice
Medical Imaging, Machine Learning
Citation (CITATION.cff)
cff-version: 1.2.0
message: "If you use this software, please cite it as below."
authors:
- family-names: "Bloice"
given-names: "Marcus D."
- family-names: "Pfeifer"
given-names: "Bastian"
title: "Pyrea"
version: 1.0.10
date-released: 2022-09-29
url: "https://github.com/mdbloice/Pyrea"
preferred-citation:
type: article
authors:
- family-names: "Pfeifer"
given-names: "Bastian"
- family-names: "Bloice"
given-names: "Marcus D."
- family-names: "Schimek"
given-names: "Michael G."
doi: "10.1016/j.jbi.2023.104406"
journal: "Journal of Biomedical Informatics"
title: "Parea: multi-view ensemble clustering for cancer subtype discovery"
year: 2023
GitHub Events
Total
- Watch event: 3
- Issue comment event: 1
- Push event: 5
- Create event: 1
Last Year
- Watch event: 3
- Issue comment event: 1
- Push event: 5
- Create event: 1
Committers
Last synced: over 1 year ago
Top Committers
| Name | Commits | |
|---|---|---|
| Marcus Bloice | m****e@m****t | 170 |
| pievos101 | b****r@g****m | 8 |
Committer Domains (Top 20 + Academic)
Issues and Pull Requests
Last synced: 6 months ago
All Time
- Total issues: 2
- Total pull requests: 0
- Average time to close issues: 9 months
- Average time to close pull requests: N/A
- Total issue authors: 2
- Total pull request authors: 0
- Average comments per issue: 2.5
- Average comments per pull request: 0
- Merged pull requests: 0
- Bot issues: 0
- Bot pull requests: 0
Past Year
- Issues: 0
- Pull requests: 0
- Average time to close issues: N/A
- Average time to close pull requests: N/A
- Issue authors: 0
- Pull request authors: 0
- Average comments per issue: 0
- Average comments per pull request: 0
- Merged pull requests: 0
- Bot issues: 0
- Bot pull requests: 0
Top Authors
Issue Authors
- yun-lai-yun-qu (1)
- PengleiGao (1)
Pull Request Authors
Top Labels
Issue Labels
Pull Request Labels
Packages
- Total packages: 1
-
Total downloads:
- pypi 63 last-month
- Total dependent packages: 0
- Total dependent repositories: 0
- Total versions: 13
- Total maintainers: 1
pypi.org: pyrea
Multi-view clustering with deep ensemble structures.
- Homepage: https://github.com/mdbloice/Pyrea
- Documentation: https://pyrea.readthedocs.io/
- License: MIT
-
Latest release: 1.0.12
published about 1 year ago
Rankings
Maintainers (1)
Dependencies
- actions/checkout v3 composite
- actions/setup-python v3 composite
- numpy >=1.0.0
- scikit-learn >=1.1.0
- numpy >1.0.0
- pandas >0.20.0
- scikit-learn >1.1.0
- scipy >1.5.4


