enzyme
High-performance automatic differentiation of LLVM and MLIR.
Science Score: 57.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
○CITATION.cff file
-
✓codemeta.json file
Found codemeta.json file -
✓.zenodo.json file
Found .zenodo.json file -
✓DOI references
Found 5 DOI reference(s) in README -
○Academic publication links
-
✓Committers with academic emails
6 of 62 committers (9.7%) from academic institutions -
✓Institutional organization owner
Organization enzymead has institutional domain (enzyme.mit.edu) -
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (14.3%) to scientific vocabulary
Keywords
Keywords from Contributors
Repository
High-performance automatic differentiation of LLVM and MLIR.
Basic Info
- Host: GitHub
- Owner: EnzymeAD
- License: other
- Language: LLVM
- Default Branch: main
- Homepage: https://enzyme.mit.edu
- Size: 630 MB
Statistics
- Stars: 1,449
- Watchers: 32
- Forks: 139
- Open Issues: 202
- Releases: 191
Topics
Metadata Files
Readme.md
 The Enzyme High-Performance Automatic Differentiator of LLVM and MLIR
The Enzyme High-Performance Automatic Differentiator of LLVM and MLIR
Enzyme is a plugin that performs automatic differentiation (AD) of statically analyzable LLVM and MLIR.
Enzyme can be used by calling __enzyme_autodiff on a function to be differentiated as shown below.
Running the Enzyme transformation pass then replaces the call to __enzyme_autodiff with the gradient of its first argument.
```c
double foo(double);
double gradfoo(double x) { return _enzyme_autodiff(foo, x); } ```
Enzyme is highly-efficient and its ability to perform AD on optimized code allows Enzyme to meet or exceed the performance of state-of-the-art AD tools.
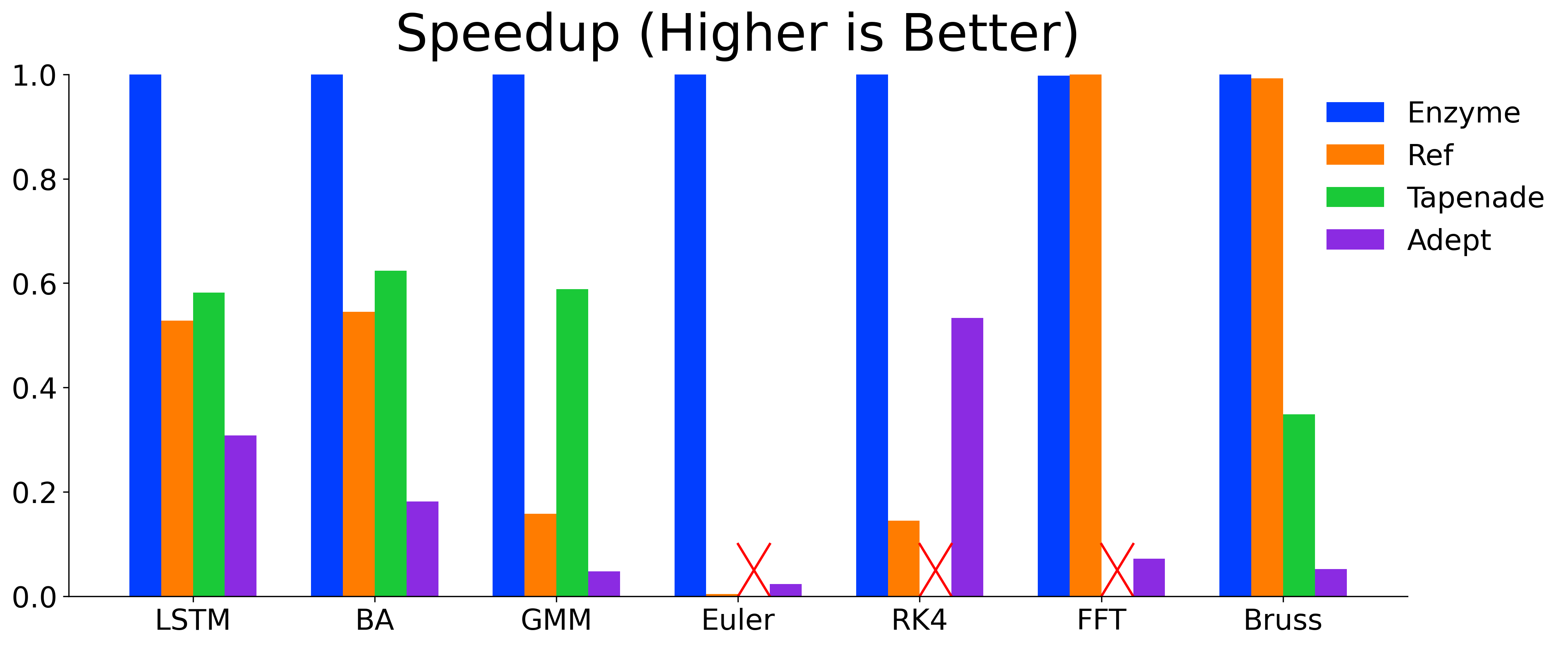
Detailed information on installing and using Enzyme can be found on our website: https://enzyme.mit.edu.
A short example of how to install Enzyme is below:
cd /path/to/Enzyme/enzyme
mkdir build && cd build
cmake -G Ninja .. -DLLVM_DIR=/path/to/llvm/lib/cmake/llvm -DLLVM_EXTERNAL_LIT=/path/to/lit/lit.py
ninja
Or, install Enzyme using a package manager:
Homebrew
brew install enzyme
Spack
spack install enzyme
Nix
nix-shell -p enzyme
To get involved or if you have questions, please join our mailing list.
If using this code in an academic setting, please cite the following three papers (first for Enzyme as a whole, second for GPU+optimizations, and third for AD of all other parallel programs (OpenMP, MPI, Julia Tasks, etc.)):
@inproceedings{NEURIPS2020_9332c513,
author = {Moses, William and Churavy, Valentin},
booktitle = {Advances in Neural Information Processing Systems},
editor = {H. Larochelle and M. Ranzato and R. Hadsell and M. F. Balcan and H. Lin},
pages = {12472--12485},
publisher = {Curran Associates, Inc.},
title = {Instead of Rewriting Foreign Code for Machine Learning, Automatically Synthesize Fast Gradients},
url = {https://proceedings.neurips.cc/paper/2020/file/9332c513ef44b682e9347822c2e457ac-Paper.pdf},
volume = {33},
year = {2020}
}
@inproceedings{10.1145/3458817.3476165,
author = {Moses, William S. and Churavy, Valentin and Paehler, Ludger and H\"{u}ckelheim, Jan and Narayanan, Sri Hari Krishna and Schanen, Michel and Doerfert, Johannes},
title = {Reverse-Mode Automatic Differentiation and Optimization of GPU Kernels via Enzyme},
year = {2021},
isbn = {9781450384421},
publisher = {Association for Computing Machinery},
address = {New York, NY, USA},
url = {https://doi.org/10.1145/3458817.3476165},
doi = {10.1145/3458817.3476165},
booktitle = {Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis},
articleno = {61},
numpages = {16},
keywords = {CUDA, LLVM, ROCm, HPC, AD, GPU, automatic differentiation},
location = {St. Louis, Missouri},
series = {SC '21}
}
@inproceedings{10.5555/3571885.3571964,
author = {Moses, William S. and Narayanan, Sri Hari Krishna and Paehler, Ludger and Churavy, Valentin and Schanen, Michel and H\"{u}ckelheim, Jan and Doerfert, Johannes and Hovland, Paul},
title = {Scalable Automatic Differentiation of Multiple Parallel Paradigms through Compiler Augmentation},
year = {2022},
isbn = {9784665454445},
publisher = {IEEE Press},
booktitle = {Proceedings of the International Conference on High Performance Computing, Networking, Storage and Analysis},
articleno = {60},
numpages = {18},
keywords = {automatic differentiation, tasks, OpenMP, compiler, Julia, parallel, Enzyme, C++, RAJA, hybrid parallelization, MPI, distributed, LLVM},
location = {Dallas, Texas},
series = {SC '22}
}
Both Julia bindings and Rust bindings are available for Enzyme.
Owner
- Name: Enzyme Automatic Differentiation Compiler
- Login: EnzymeAD
- Kind: organization
- Email: enzyme-dev@googlegroups.com
- Location: United States of America
- Website: https://enzyme.mit.edu
- Repositories: 13
- Profile: https://github.com/EnzymeAD
GitHub Events
Total
- Fork event: 23
- Create event: 216
- Commit comment event: 1
- Issues event: 82
- Release event: 37
- Watch event: 156
- Delete event: 122
- Member event: 3
- Issue comment event: 214
- Push event: 842
- Pull request review event: 230
- Pull request review comment event: 165
- Pull request event: 400
Last Year
- Fork event: 23
- Create event: 216
- Commit comment event: 1
- Issues event: 82
- Release event: 37
- Watch event: 156
- Delete event: 122
- Member event: 3
- Issue comment event: 214
- Push event: 842
- Pull request review event: 230
- Pull request review comment event: 165
- Pull request event: 400
Committers
Last synced: 10 months ago
Top Committers
| Name | Commits | |
|---|---|---|
| William Moses | gh@w****m | 1,809 |
| Tim Gymnich | t****h@m****m | 207 |
| Manuel Drehwald | g****t@m****o | 95 |
| Valentin Churavy | v****y@g****m | 49 |
| Ludger Paehler | l****r@t****e | 34 |
| Oleksandr "Alex" Zinenko | z****o@g****m | 17 |
| Paul Berg | n****b@g****m | 10 |
| Tim Kaler | t****k@m****u | 8 |
| Jacob Mai Peng | j****g@g****m | 8 |
| Ivan R. Ivanov | i****a@m****p | 8 |
| Brant Qian | s****q@g****m | 7 |
| dependabot[bot] | 4****] | 7 |
| Ubuntu | u****u@i****l | 6 |
| Ubuntu | u****u@i****l | 6 |
| dan | t****2@g****m | 5 |
| Jesse Michel | j****l@m****u | 5 |
| Matt Bolitho | m****e@g****m | 5 |
| jumerckx | 3****x | 5 |
| Mo Tabbara | 1****a | 3 |
| Sam | s****h | 3 |
| Romain Moyard | r****d@g****m | 3 |
| Max Aehle | m****e@s****e | 3 |
| Martin Robinson | m****s@g****m | 3 |
| Carlo Cabrera | 3****b | 3 |
| Jordan Rupprecht | j****t@g****m | 2 |
| min xu | 1****s | 2 |
| Pratyush Das | r****s@g****m | 2 |
| Nicholas Wilson | t****r | 2 |
| Nestor Demeure | n****e@g****m | 2 |
| Michel Schanen | m****n@g****m | 2 |
| and 32 more... | ||
Committer Domains (Top 20 + Academic)
Issues and Pull Requests
Last synced: 6 months ago
All Time
- Total issues: 146
- Total pull requests: 910
- Average time to close issues: 3 months
- Average time to close pull requests: 22 days
- Total issue authors: 66
- Total pull request authors: 44
- Average comments per issue: 1.76
- Average comments per pull request: 0.44
- Merged pull requests: 703
- Bot issues: 0
- Bot pull requests: 15
Past Year
- Issues: 67
- Pull requests: 434
- Average time to close issues: 8 days
- Average time to close pull requests: 3 days
- Issue authors: 37
- Pull request authors: 29
- Average comments per issue: 0.81
- Average comments per pull request: 0.33
- Merged pull requests: 338
- Bot issues: 0
- Bot pull requests: 7
Top Authors
Issue Authors
- ZuseZ4 (24)
- wsmoses (18)
- davidedelvento (8)
- minansys (6)
- dime10 (4)
- ipcamit (4)
- jandrej (4)
- alecjacobson (4)
- ax3l (3)
- GregTheMadMonk (2)
- cgeoga (2)
- spookyGh0st (2)
- shadow-orange41 (2)
- samuelpmish (2)
- RasmitDevkota (2)
Pull Request Authors
- wsmoses (611)
- ZuseZ4 (68)
- tgymnich (33)
- Pangoraw (32)
- ivanradanov (15)
- dependabot[bot] (15)
- ftynse (13)
- jumerckx (12)
- MattBolitho (12)
- vimarsh6739 (9)
- pengmai (9)
- martinjrobins (6)
- Brant-Skywalker (6)
- avik-pal (6)
- rmoyard (5)
Top Labels
Issue Labels
Pull Request Labels
Dependencies
- actions/checkout v3 composite
- actions/checkout v1 composite
- actions/checkout v3 composite
- actions/checkout v3 composite
- mattnotmitt/doxygen-action v1.9.2 composite
- wsmoses/action-deploy-scripts main composite
- actions/checkout v3 composite
- actions/checkout v3 composite
- julia-actions/julia-buildpkg v1 composite
- julia-actions/julia-runtest v1 composite
- julia-actions/setup-julia v1 composite
- actions/cache v2 composite
- actions/checkout v3 composite
- DoozyX/clang-format-lint-action v0.11 composite
- actions/checkout v3 composite
- actions/checkout v3 composite
- peter-evans/create-pull-request v3 composite
- tibdex/github-app-token v1 composite
- actions/checkout v3 composite
- actions/checkout v3 composite