concise-concepts
This repository contains an easy and intuitive approach to few-shot NER using most similar expansion over spaCy embeddings. Now with entity scoring.
Science Score: 44.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
✓CITATION.cff file
Found CITATION.cff file -
✓codemeta.json file
Found codemeta.json file -
✓.zenodo.json file
Found .zenodo.json file -
○DOI references
-
○Academic publication links
-
○Committers with academic emails
-
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (13.2%) to scientific vocabulary
Keywords
Repository
This repository contains an easy and intuitive approach to few-shot NER using most similar expansion over spaCy embeddings. Now with entity scoring.
Basic Info
Statistics
- Stars: 244
- Watchers: 6
- Forks: 14
- Open Issues: 5
- Releases: 22
Topics
Metadata Files
README.md
Concise Concepts
When wanting to apply NER to concise concepts, it is really easy to come up with examples, but pretty difficult to train an entire pipeline. Concise Concepts uses few-shot NER based on word embedding similarity to get you going with easy! Now with entity scoring!
Usage
This library defines matching patterns based on the most similar words found in each group, which are used to fill a spaCy EntityRuler. To better understand the rule definition, I recommend playing around with the spaCy Rule-based Matcher Explorer.
Tutorials
TechVizTheDataScienceGuy created a nice tutorial on how to use it.
The section Matching Pattern Rules expands on the construction, analysis and customization of these matching patterns.
Install
pip install concise-concepts
Quickstart
Take a look at the configuration section for more info.
Spacy Pipeline Component
Note that, custom embedding models are passed via model_path.
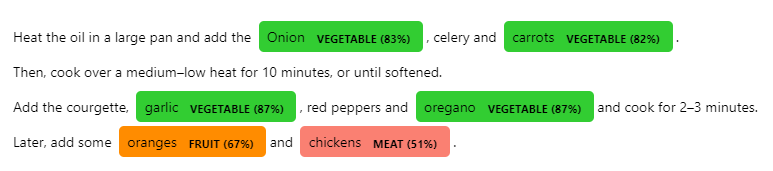
```python import spacy from spacy import displacy
data = { "fruit": ["apple", "pear", "orange"], "vegetable": ["broccoli", "spinach", "tomato"], "meat": ['beef', 'pork', 'turkey', 'duck'] }
text = """ Heat the oil in a large pan and add the Onion, celery and carrots. Then, cook over a medium–low heat for 10 minutes, or until softened. Add the courgette, garlic, red peppers and oregano and cook for 2–3 minutes. Later, add some oranges and chickens. """
nlp = spacy.load("encoreweb_md", disable=["ner"])
nlp.addpipe( "conciseconcepts", config={ "data": data, "entscore": True, # Entity Scoring section "verbose": True, "excludepos": ["VERB", "AUX"], "excludedep": ["DOBJ", "PCOMP"], "includecompoundwords": False, "jsonpath": "./fruitful_patterns.json", "topn": (100,500,300) }, ) doc = nlp(text)
options = { "colors": {"fruit": "darkorange", "vegetable": "limegreen", "meat": "salmon"}, "ents": ["fruit", "vegetable", "meat"], }
ents = doc.ents for ent in ents: newlabel = f"{ent.label} ({ent..entscore:.0%})" options["colors"][newlabel] = options["colors"].get(ent.label.lower(), None) options["ents"].append(newlabel) ent.label = new_label doc.ents = ents
displacy.render(doc, style="ent", options=options)
```
Standalone
This might be useful when iterating over few_shot training data when not wanting to reload larger models continuously.
Note that, custom embedding models are passed via model.
```python import gensim import spacy
from concise_concepts import Conceptualizer
model = gensim.downloader.load("fasttext-wiki-news-subwords-300") nlp = spacy.load("encoreweb_sm") data = { "disease": ["cancer", "diabetes", "heart disease", "influenza", "pneumonia"], "symptom": ["headache", "fever", "cough", "nausea", "vomiting", "diarrhea"], } conceptualizer = Conceptualizer(nlp, data, model) conceptualizer.nlp("I have a headache and a fever.").ents
data = { "disease": ["cancer", "diabetes"], "symptom": ["headache", "fever"], } conceptualizer = Conceptualizer(nlp, data, model) conceptualizer.nlp("I have a headache and a fever.").ents ```
Configuration
Matching Pattern Rules
A general introduction about the usage of matching patterns in the usage section.
Customizing Matching Pattern Rules
Even though the baseline parameters provide a decent result, the construction of these matching rules can be customized via the config passed to the spaCy pipeline.
exclude_pos: A list of POS tags to be excluded from the rule-based match.exclude_dep: A list of dependencies to be excluded from the rule-based match.include_compound_words: If True, it will include compound words in the entity. For example, if the entity is "New York", it will also include "New York City" as an entity.case_sensitive: Whether to match the case of the words in the text.
Analyze Matching Pattern Rules
To motivate actually looking at the data and support interpretability, the matching patterns that have been generated are stored as ./main_patterns.json. This behavior can be changed by using the json_path variable via the config passed to the spaCy pipeline.
Fuzzy matching using spaczz
fuzzy: A boolean value that determines whether to use fuzzy matching
```python data = { "fruit": ["apple", "pear", "orange"], "vegetable": ["broccoli", "spinach", "tomato"], "meat": ["beef", "pork", "fish", "lamb"] }
nlp.addpipe("conciseconcepts", config={"data": data, "fuzzy": True}) ```
Most Similar Word Expansion
topn: Use a specific number of words to expand over.
```python data = { "fruit": ["apple", "pear", "orange"], "vegetable": ["broccoli", "spinach", "tomato"], "meat": ["beef", "pork", "fish", "lamb"] }
topn = [50, 50, 150]
assert len(topn) == len
nlp.addpipe("conciseconcepts", config={"data": data, "topn": topn}) ```
Entity Scoring
ent_score: Use embedding based word similarity to score entities against their groups
```python import spacy
data = { "ORG": ["Google", "Apple", "Amazon"], "GPE": ["Netherlands", "France", "China"], }
text = """Sony was founded in Japan."""
nlp = spacy.load("encoreweblg") nlp.addpipe("conciseconcepts", config={"data": data, "entscore": True, "case_sensitive": True}) doc = nlp(text)
print([(ent.text, ent.label, ent..ent_score) for ent in doc.ents])
output
[('Sony', 'ORG', 0.5207586), ('Japan', 'GPE', 0.7371268)]
```
Custom Embedding Models
model_path: Use customsense2vec.Sense2Vec,gensim.Word2vecgensim.FastText, orgensim.KeyedVectors, or a pretrained model from gensim library or a custom model path. For using asense2vec.Sense2Vectake a look here.model: within standalone usage, it is possible to pass these models directly.
```python data = { "fruit": ["apple", "pear", "orange"], "vegetable": ["broccoli", "spinach", "tomato"], "meat": ["beef", "pork", "fish", "lamb"] }
model from https://radimrehurek.com/gensim/downloader.html or path to local file
model_path = "glove-wiki-gigaword-300"
nlp.addpipe("conciseconcepts", config={"data": data, "modelpath": modelpath}) ````
Owner
- Name: David Berenstein
- Login: davidberenstein1957
- Kind: user
- Location: Madrid
- Company: @argilla-io
- Website: https://www.linkedin.com/in/david-berenstein-1bab11105/
- Repositories: 2
- Profile: https://github.com/davidberenstein1957
👨🏽🍳 Cooking, 👨🏽💻 Coding, 🏆 Committing Developer Advocate @argilla-io
Citation (CITATION.cff)
cff-version: 1.0.0
message: "If you use this software, please cite it as below."
authors:
- family-names: David
given-names: Berenstein
title: "Concise Concepts - an easy and intuitive approach to few-shot NER using most similar expansion over spaCy embeddings."
version: 0.7.3
date-released: 2022-12-31
GitHub Events
Total
- Watch event: 7
Last Year
- Watch event: 7
Committers
Last synced: 11 months ago
Top Committers
| Name | Commits | |
|---|---|---|
| david | d****n@g****m | 58 |
| David Berenstein | d****n@p****m | 29 |
| Tom Aarsen | C****v@g****m | 2 |
| simon | i****o@d****g | 2 |
| vincent d warmerdam | v****m@g****m | 1 |
| Robin de Heer | r****r@p****m | 1 |
Committer Domains (Top 20 + Academic)
Issues and Pull Requests
Last synced: 6 months ago
All Time
- Total issues: 34
- Total pull requests: 13
- Average time to close issues: about 1 month
- Average time to close pull requests: 1 day
- Total issue authors: 17
- Total pull request authors: 6
- Average comments per issue: 3.24
- Average comments per pull request: 0.08
- Merged pull requests: 11
- Bot issues: 0
- Bot pull requests: 0
Past Year
- Issues: 0
- Pull requests: 0
- Average time to close issues: N/A
- Average time to close pull requests: N/A
- Issue authors: 0
- Pull request authors: 0
- Average comments per issue: 0
- Average comments per pull request: 0
- Merged pull requests: 0
- Bot issues: 0
- Bot pull requests: 0
Top Authors
Issue Authors
- nv78 (1)
Pull Request Authors
- davidberenstein1957 (1)
Top Labels
Issue Labels
Pull Request Labels
Dependencies
- atomicwrites 1.4.0 develop
- attrs 21.4.0 develop
- iniconfig 1.1.1 develop
- pluggy 1.0.0 develop
- py 1.11.0 develop
- pytest 7.1.0 develop
- tomli 2.0.1 develop
- blis 0.7.6
- catalogue 2.0.6
- certifi 2021.10.8
- charset-normalizer 2.0.12
- click 7.1.2
- colorama 0.4.4
- cymem 2.0.6
- gensim 4.1.2
- idna 3.3
- importlib-metadata 4.11.3
- jinja2 3.0.3
- markupsafe 2.1.0
- murmurhash 1.0.6
- numpy 1.22.3
- packaging 21.3
- pathy 0.6.1
- preshed 3.0.6
- pydantic 1.7.4
- pyparsing 3.0.7
- requests 2.27.1
- scipy 1.6.1
- smart-open 5.2.1
- spacy 3.0.0
- spacy-legacy 3.0.9
- srsly 2.4.2
- thinc 8.0.13
- tqdm 4.63.0
- typer 0.3.2
- typing-extensions 3.10.0.2
- urllib3 1.26.8
- wasabi 0.9.0
- zipp 3.7.0
- black ^22.3.0 develop
- flake8 ^4.0.1 develop
- flake8-bugbear ^22.3.23 develop
- flake8-docstrings ^1.6.0 develop
- isort ^5.10.1 develop
- pep8-naming ^0.12.1 develop
- pre-commit ^2.17.0 develop
- pytest ^7.0.1 develop
- gensim ^4
- python ^3.7
- spacy ^3
- actions/checkout v3 composite
- actions/setup-python v3 composite
- actions/checkout v3 composite
- actions/setup-python v3 composite
- pypa/gh-action-pypi-publish 27b31702a0e7fc50959f5ad993c78deac1bdfc29 composite



