vedo
A python module for scientific analysis of 3D data based on VTK and Numpy
Science Score: 59.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
○CITATION.cff file
-
✓codemeta.json file
Found codemeta.json file -
✓.zenodo.json file
Found .zenodo.json file -
✓DOI references
Found 33 DOI reference(s) in README -
✓Academic publication links
Links to: frontiersin.org, acm.org, aps.org, zenodo.org -
✓Committers with academic emails
2 of 45 committers (4.4%) from academic institutions -
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (15.8%) to scientific vocabulary
Keywords
Keywords from Contributors
Repository
A python module for scientific analysis of 3D data based on VTK and Numpy
Basic Info
- Host: GitHub
- Owner: marcomusy
- License: mit
- Language: Python
- Default Branch: master
- Homepage: https://vedo.embl.es
- Size: 87.4 MB
Statistics
- Stars: 2,193
- Watchers: 32
- Forks: 273
- Open Issues: 159
- Releases: 58
Topics
Metadata Files
README.md

Your friendly python module
for scientific analysis and visualization of 3d objects.
💾 Installation
bash
pip install vedo
additional installation details [click to expand]
- To install the latest _dev_ version of `vedo`: ```bash pip install -U git+https://github.com/marcomusy/vedo.git ``` - To install from the conda-forge channel: ```bash conda install -c conda-forge vedo ```📙 Documentation
The webpage of the library with documentation is available here.
📌 Need help? Have a question, or wish to ask for a missing feature? Do not hesitate to ask any questions on the image.sc forum or by opening a github issue.
🎨 Features
The library includes a large set of working examples for a wide range of functionalities
working with polygonal meshes and point clouds [click to expand]
- Import meshes from VTK format, STL, Wavefront OBJ, 3DS, Dolfin-XML, Neutral, GMSH, OFF, PCD (PointCloud), - Export meshes as ASCII or binary to VTK, STL, OBJ, PLY ... formats. - Analysis tools like Moving Least Squares, mesh morphing and more.. - Tools to visualize and edit meshes (cutting a mesh with another mesh, slicing, normalizing, moving vertex positions, etc..). - Split mesh based on surface connectivity. Extract the largest connected area. - Calculate areas, volumes, center of mass, average sizes etc. - Calculate vertex and face normals, curvatures, feature edges. Fill mesh holes. - Subdivide faces of a mesh, increasing the number of vertex points. Mesh simplification. - Coloring and thresholding of meshes based on associated scalar or vectorial data. - Point-surface operations: find nearest points, determine if a point lies inside or outside of a mesh. - Create primitive shapes: spheres, arrows, cubes, torus, ellipsoids... - Generate glyphs (associate a mesh to every vertex of a source mesh). - Create animations easily by just setting the position of the displayed objects in the 3D scene. Add trailing lines and shadows to moving objects is supported. - Straightforward support for multiple sync-ed or independent renderers in the same window. - Registration (alignment) of meshes with different techniques. - Mesh smoothing. - Delaunay triangulation in 2D and 3D. - Generate meshes by joining nearby lines in space. - Find the closest path from one point to another, traveling along the edges of a mesh. - Find the intersection of a mesh with lines, planes or other meshes. - Interpolate scalar and vectorial fields with Radial Basis Functions and Thin Plate Splines. - Add sliders and buttons to interact with the scene and the individual objects. - Visualization of tensors. - Analysis of Point Clouds - Moving Least Squares smoothing of 2D, 3D and 4D clouds - Fit lines, planes, spheres and ellipsoids in space - Identify outliers in a distribution of points - Decimate a cloud to a uniform distribution.working with volumetric data and tetrahedral meshes
- Import data from VTK format volumetric TIFF stacks, DICOM, SLC, MHD and more - Import 2D images as PNG, JPEG, BMP - Isosurfacing of volumes - Composite and maximum projection volumetric rendering - Generate volumetric signed-distance data from an input surface mesh - Probe volumes with lines and planes - Generate stream-lines and stream-tubes from vectorial fields - Slice and crop volumes - Support for other volumetric structures (structured and grid data)plotting and histogramming in 2D and 3D
- Polygonal 3D text rendering with Latex-like syntax and unicode characters, with 30 different fonts. - Fully customizable axis styles - donut plots and pie charts - Scatter plots in 2D and 3D - Surface function plotting - 1D customizable histograms - 2D hexagonal histograms - Polar plots, spherical plots and histogramming - Draw latex-formatted formulas in the rendering window. - Quiver, violin, whisker and stream-line plots - Graphical markers analogous to matplotlibintegration with other libraries
- Integration with the [Qt5](https://www.qt.io/) framework. - Support for [FEniCS/Dolfin](https://fenicsproject.org/) platform for visualization of PDE/FEM solutions. - Interoperability with the [trimesh](https://trimsh.org/), [pyvista](https://github.com/pyvista/pyvista) and [pymeshlab](https://github.com/cnr-isti-vclab/PyMeshLab) libraries. - Export 3D scenes and embed them into a [web page](https://vedo.embl.es/examples/fenics_elasticity.html). - Embed 3D scenes in *jupyter* notebooks with [K3D](https://github.com/K3D-tools/K3D-jupyter) (can export an interactive 3D-snapshot page [here](https://vedo.embl.es/examples/geo_scene.html)).⌨ Command Line Interface
Visualize a polygonal mesh or a volume from a terminal window simply with:
bash
vedo https://vedo.embl.es/examples/data/embryo.tif
volumetric files (slc, tiff, DICOM...) can be visualized in different modes [click to expand]
|Volume 3D slicing`vedo --slicer embryo.slc`| Ray-casting
`vedo -g`| 2D slicing
`vedo --slicer2d`| |:--------|:-----|:--------| | 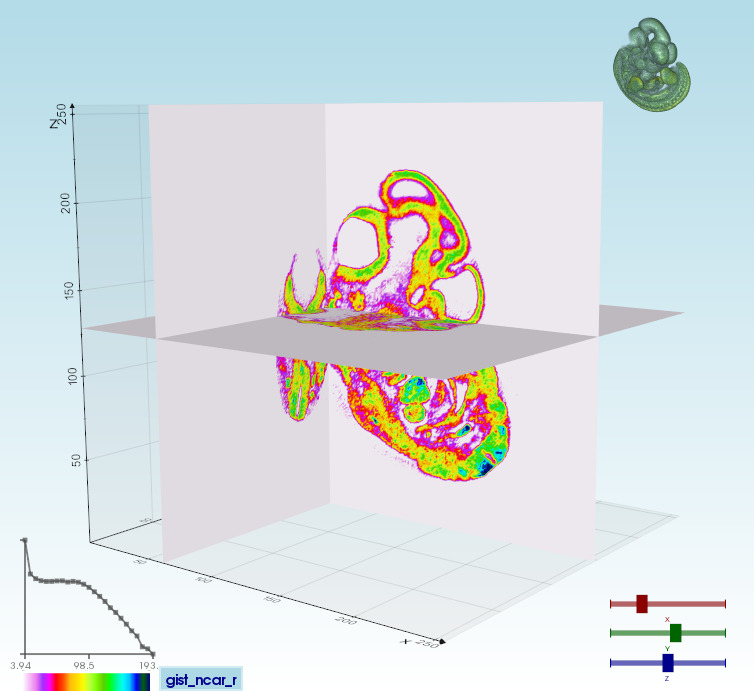 |  |  |
Type vedo -h for the complete list of options.
🐾 Gallery
vedo currently includes 300+ working examples and notebooks.
Run any of the built-in examples. In a terminal type: vedo -r warp2
Check out the example galleries organized by subject here:

✏ Contributing
Any contributions are greatly appreciated! If you have a suggestion that would make this better, please fork the repo and create a pull request. This is how: ```bash
1. Fork the repository on GitHub then clone your fork locally:
git clone https://github.com/your-username/vedo.git
2. Create a new branch for your feature or bugfix:
git checkout -b feature/my-feature
3. Make your changes and commit them:
git commit -m "Description of my feature"
4. Push your changes to your fork:
git push origin feature/my-feature
5. Open a Pull Request on the main repository.
``` You can also simply open an issue with the tag "enhancement".
📜 References
Scientific publications leveraging vedo:
- X. Diego et al.: "Key features of Turing systems are determined purely by network topology", Phys. Rev. X 8, 021071, DOI.
- M. Musy, K. Flaherty et al.: "A Quantitative Method for Staging Mouse Limb Embryos based on Limb Morphometry", Development (2018) 145 (7): dev154856, DOI.
- F. Claudi, A. L. Tyson, T. Branco, "Brainrender. A python based software for visualisation of neuroanatomical and morphological data.", eLife 2021;10:e65751, DOI.
- J. S. Bennett, D. Sijacki, "Resolving shocks and filaments in galaxy formation simulations: effects on gas properties and star formation in the circumgalactic medium", Monthly Notices of the Royal Astronomical Society, Volume 499, Issue 1, DOI.
- J.D.P. Deshapriya et al., "Spectral analysis of craters on (101955) Bennu". Icarus 2020, DOI.
- A. Pollack et al., "Stochastic inversion of gravity, magnetic, tracer, lithology, and fault data for geologically realistic structural models: Patua Geothermal Field case study", Geothermics, Volume 95, September 2021, DOI.
- X. Lu et al., "3D electromagnetic modeling of graphitic faults in the Athabasca Basin using a finite-volume time-domain approach with unstructured grids", Geophysics, DOI.
- M. Deepa Maheshvare et al., "A Graph-Based Framework for Multiscale Modeling of Physiological Transport", Front. Netw. Physiol. 1:802881, DOI.
- F. Claudi, T. Branco, "Differential geometry methods for constructing manifold-targeted recurrent neural networks", bioRxiv 2021.10.07.463479, DOI.
- J. Klatzow, G. Dalmasso, N. Martínez-Abadías, J. Sharpe, V. Uhlmann, "µMatch: 3D shape correspondence for microscopy data", Front. Comput. Sci., 15 February 2022. DOI
- G. Dalmasso et al., "4D reconstruction of murine developmental trajectories using spherical harmonics", Developmental Cell 57, 1–11 September 2022, DOI.
- D.J.E Waibel et al., "Capturing Shape Information with Multi-scale Topological Loss Terms for 3D Reconstruction", Lecture Notes in Computer Science, vol 13434. Springer, Cham. DOI.
- N. Lamb et al., "DeepJoin: Learning a Joint Occupancy, Signed Distance, and Normal Field Function for Shape Repair", ACM Transactions on Graphics (TOG), vol 41, 6, 2022. DOI
- J. Cotterell et al., "Cell 3D Positioning by Optical encoding (C3PO) and its application to spatial transcriptomics", bioRxiv 2024.03.12.584578. DOI
- L. Aviñó-Esteban et al., "Spatio-temporal reconstruction of gene expression patterns in developing mice", Development (2025) 152. DOI
- J.S. Posada et al., "morphoHeart: A quantitative tool for integrated 3D morphometric analyses of heart and ECM during embryonic development", PLOS Biology 23(1). DOI
Have you found this software useful for your research? Star ✨ the project and cite it as:
M. Musy et al.,
"vedo, a python module for scientific analysis and visualization of 3D objects and point clouds",
Zenodo, 2021, doi: 10.5281/zenodo.7019968.
Owner
- Name: Marco Musy
- Login: marcomusy
- Kind: user
- Location: Barcelona
- Company: EMBL
- Website: https://es.linkedin.com/in/marco-musy
- Repositories: 16
- Profile: https://github.com/marcomusy
EMBL
GitHub Events
Total
- Create event: 2
- Release event: 1
- Issues event: 92
- Watch event: 148
- Issue comment event: 168
- Push event: 81
- Pull request review event: 2
- Pull request event: 17
- Fork event: 12
Last Year
- Create event: 2
- Release event: 1
- Issues event: 92
- Watch event: 148
- Issue comment event: 168
- Push event: 81
- Pull request review event: 2
- Pull request event: 17
- Fork event: 12
Committers
Last synced: 9 months ago
Top Committers
| Name | Commits | |
|---|---|---|
| marcomusy | m****y@g****m | 1,608 |
| Guillaume Jacquenot | g****t@g****m | 82 |
| Jeffrey Wardman | j****w@z****m | 11 |
| Giovanni Dalmasso | g****a | 11 |
| Marco Musy | m****y@M****l | 10 |
| Jaewook Lee | j****2@g****m | 10 |
| Johannes Müller | 3****r | 7 |
| Louis-Pujol | l****l@l****t | 6 |
| Ruben de Bruin | 3****n | 6 |
| neoglez | s****2@s****t | 5 |
| Zhi-Qiang Zhou @ MacBookPro16,1 | z****u@o****m | 5 |
| Mikael Tulldahl | m****l@l****e | 4 |
| Poisonous | 4****d | 3 |
| robin | r****t@g****m | 3 |
| Brian Denheyer | b****d@a****m | 2 |
| Ahinoam Pollack | a****p@g****m | 2 |
| Antony Lee | a****e@g****m | 2 |
| Brandon Hacha | 2****a | 2 |
| Eden.Kellner | e****l@g****m | 2 |
| FedeClaudi | f****i@p****m | 2 |
| Justin Kunimune | j****e@g****m | 2 |
| The Codacy Badger | b****r@c****m | 2 |
| Umut E. S. | u****l@g****m | 2 |
| Xushan Lu | l****n@g****m | 2 |
| icemtel | i****l@g****m | 2 |
| ilorevilo | 4****o | 1 |
| daizhirui | d****i@h****m | 1 |
| RichardScottOZ | 7****Z | 1 |
| Peter Mitrano | m****r@g****m | 1 |
| Diego Tavares Volpatto | d****o@e****o | 1 |
| and 15 more... | ||
Committer Domains (Top 20 + Academic)
Issues and Pull Requests
Last synced: 6 months ago
All Time
- Total issues: 380
- Total pull requests: 63
- Average time to close issues: about 2 months
- Average time to close pull requests: 10 days
- Total issue authors: 160
- Total pull request authors: 23
- Average comments per issue: 3.24
- Average comments per pull request: 0.92
- Merged pull requests: 51
- Bot issues: 0
- Bot pull requests: 0
Past Year
- Issues: 59
- Pull requests: 16
- Average time to close issues: 15 days
- Average time to close pull requests: 2 days
- Issue authors: 39
- Pull request authors: 5
- Average comments per issue: 1.81
- Average comments per pull request: 1.06
- Merged pull requests: 15
- Bot issues: 0
- Bot pull requests: 0
Top Authors
Issue Authors
- JeffreyWardman (41)
- zhang-qiang-github (22)
- OhmPuchiss (14)
- smoothumut (12)
- ManuGraiph (10)
- LogWell (9)
- DeepaMahm (8)
- marcomusy (8)
- jo-mueller (7)
- paul0noah (6)
- ivishalanand (6)
- ttsesm (6)
- priyabiswas12 (5)
- IsabellaPa (5)
- natabma (5)
Pull Request Authors
- JeffreyWardman (12)
- marcomusy (11)
- j042 (6)
- Poisoned (6)
- Louis-Pujol (5)
- CorpsSansOrganes (5)
- zhouzq-thu (4)
- anntzer (4)
- jo-mueller (3)
- smoothumut (3)
- jkunimune (3)
- paulbrodersen (2)
- Linus-Foley (2)
- dbotwinick (2)
- PinkMushroom (2)
Top Labels
Issue Labels
Pull Request Labels
Packages
- Total packages: 3
-
Total downloads:
- pypi 46,043 last-month
- Total docker downloads: 52
-
Total dependent packages: 30
(may contain duplicates) -
Total dependent repositories: 59
(may contain duplicates) - Total versions: 68
- Total maintainers: 1
pypi.org: vedo
A python module for scientific visualization, analysis of 3D objects and point clouds.
- Homepage: https://github.com/marcomusy/vedo
- Documentation: https://vedo.embl.es
- License: MIT License Copyright (c) 2017 Marco Musy Permission is hereby granted, free of charge, to any person obtaining a copy of this software and associated documentation files (the "Software"), to deal in the Software without restriction, including without limitation the rights to use, copy, modify, merge, publish, distribute, sublicense, and/or sell copies of the Software, and to permit persons to whom the Software is furnished to do so, subject to the following conditions: The above copyright notice and this permission notice shall be included in all copies or substantial portions of the Software. THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.
-
Latest release: 2025.5.4
published 9 months ago
Rankings
Maintainers (1)
proxy.golang.org: github.com/marcomusy/vedo
- Documentation: https://pkg.go.dev/github.com/marcomusy/vedo#section-documentation
- License: mit
-
Latest release: v2025.5.4+incompatible
published 9 months ago
Rankings
conda-forge.org: vedo
- Homepage: https://github.com/marcomusy/vedo/
- License: MIT
-
Latest release: 2022.4.1
published over 3 years ago
Rankings
Dependencies
- Pygments *
- vtk *
- vtk *


