Science Score: 54.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
○CITATION.cff file
-
✓codemeta.json file
Found codemeta.json file -
○.zenodo.json file
-
✓DOI references
Found 2 DOI reference(s) in README -
✓Academic publication links
Links to: biorxiv.org, nature.com, plos.org -
✓Committers with academic emails
5 of 23 committers (21.7%) from academic institutions -
✓Institutional organization owner
Organization vanvalenlab has institutional domain (vanvalen.caltech.edu) -
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (16.0%) to scientific vocabulary
Keywords
Keywords from Contributors
Repository
Deep Learning Library for Single Cell Analysis
Basic Info
- Host: GitHub
- Owner: vanvalenlab
- License: other
- Language: Python
- Default Branch: master
- Homepage: https://deepcell.readthedocs.io
- Size: 341 MB
Statistics
- Stars: 462
- Watchers: 15
- Forks: 105
- Open Issues: 52
- Releases: 39
Topics
Metadata Files
README.md
deepcell-tf is a deep learning library for single-cell analysis of biological images. It is written in Python and built using TensorFlow 2.
This library allows users to apply pre-existing models to imaging data as well as to develop new deep learning models for single-cell analysis. This library specializes in models for cell segmentation (whole-cell and nuclear) in 2D and 3D images as well as cell tracking in 2D time-lapse datasets. These models are applicable to data ranging from multiplexed images of tissues to dynamic live-cell imaging movies.
deepcell-tf is one of several resources created by the Van Valen lab to facilitate the development and application of new deep learning methods to biology. Other projects within our DeepCell ecosystem include the DeepCell Toolbox for pre and post-processing the outputs of deep learning models, DeepCell Tracking for creating cell lineages with deep-learning-based tracking models, and the DeepCell Kiosk for deploying workflows on large datasets in the cloud. Additionally, we have developed DeepCell Label for annotating high-dimensional biological images to use as training data.
Read the documentation at deepcell.readthedocs.io.
For more information on deploying models in the cloud refer to the the Kiosk documentation.
Examples
| Raw Image | Tracked Image |
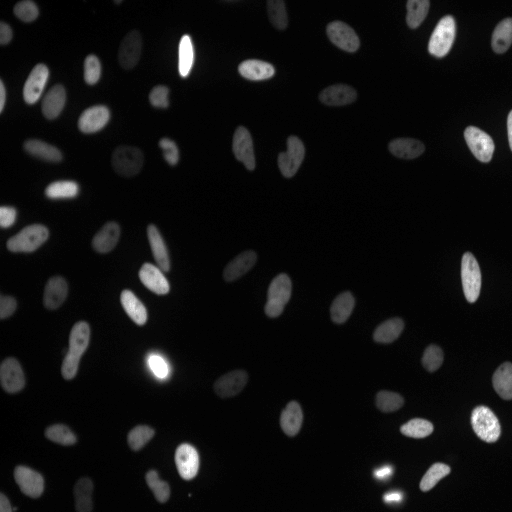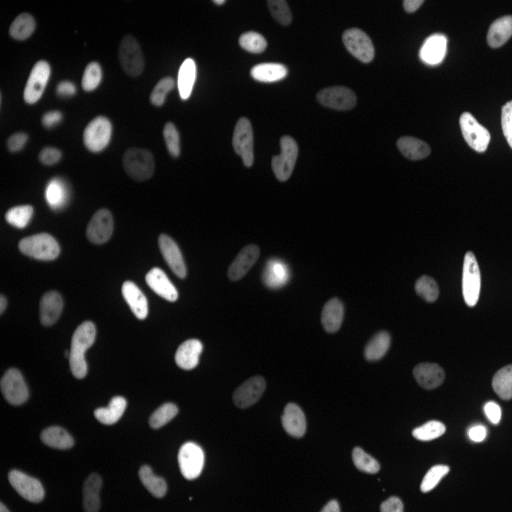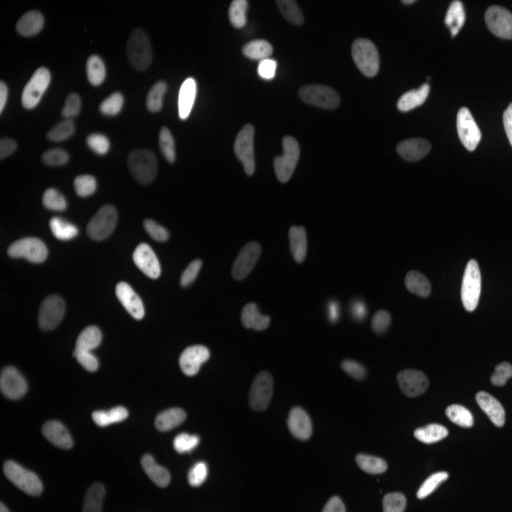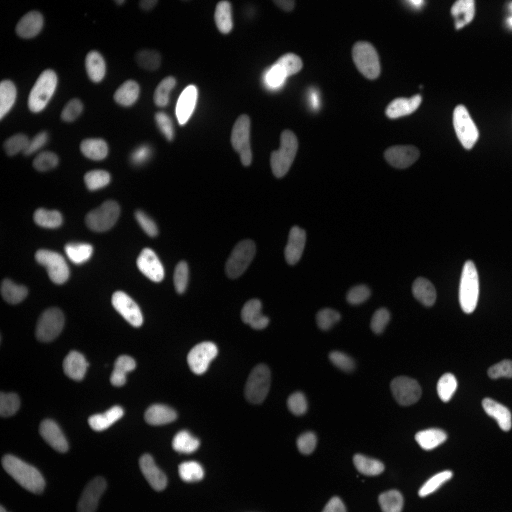
|

|
Getting Started
Install with pip
The fastest way to get started with deepcell-tf is to install the package with pip:
bash
pip install deepcell
Install with Docker
There are also docker containers with GPU support available on DockerHub. To run the library locally on a GPU, make sure you have CUDA and Docker v19.03 or later installed. For prior Docker versions, use nvidia-docker. Alternatively, Google Cloud Platform (GCP) offers prebuilt virtual machines preinstalled with CUDA, Docker, and the NVIDIA Container Toolkit.
Once docker is installed, run the following command:
```bash
Start a GPU enabled container on one GPUs
docker run --gpus '"device=0"' -it --rm \ -p 8888:8888 \ -v $PWD/notebooks:/notebooks \ -v $PWD/data:/data \ vanvalenlab/deepcell-tf:latest-gpu ```
This will start a Docker container with deepcell-tf installed and start a jupyter session using the default port 8888. This command also mounts a data folder ($PWD/data) and a notebooks folder ($PWD/notebooks) to the docker container so it can access data and Juyter notebooks stored on the host workstation. Data and models must be saved in these mounted directories to persist them outside of the running container. The default port can be changed to any non-reserved port by updating -p 8888:8888 to, e.g., -p 8080:8888. If you run across any errors getting started, you should either refer to the deepcell-tf for developers section or raise an issue on GitHub.
For examples of how to train models with the deepcell-tf library, check out the following notebooks:
DeepCell Applications and DeepCell Datasets
deepcell-tf contains two modules that greatly simplify the development and usage of deep learning models for single cell analysis. The first is deepcell.datasets, a collection of biological images that have single-cell annotations. These data include live-cell imaging movies of fluorescent nuclei (approximately 10,000 single-cell trajectories over 30 frames), as well as static images of whole cells (both phase and fluorescence images - approximately 75,000 single cell annotations). The second is deepcell.applications, which contains pre-trained models (fluorescent nuclear and phase/fluorescent whole cell) for single-cell analysis. Provided data is scaled so that the physical size of each pixel matches that in the training dataset, these models can be used out of the box on live-cell imaging data. We are currently working to expand these modules to include data and models for tissue images. Please note that they may be spun off into their own GitHub repositories in the near future.
DeepCell-tf for Developers
deepcell-tf uses docker and tensorflow to enable GPU processing. If using GCP, there are pre-built images which come with CUDA and Docker pre-installed. Otherwise, you will need to install docker and CUDA separately.
Build a local docker container, specifying the tensorflow version with TF_VERSION
bash
git clone https://github.com/vanvalenlab/deepcell-tf.git
cd deepcell-tf
docker build --build-arg TF_VERSION=2.8.0-gpu -t $USER/deepcell-tf .
Run the new docker image
```bash
'"device=0"' refers to the specific GPU(s) to run DeepCell-tf on, and is not required
docker run --gpus '"device=0"' -it \ -p 8888:8888 \ $USER/deepcell-tf:latest-gpu ```
It can also be helpful to mount the local copy of the repository and the notebooks to speed up local development. However, if you are going to mount a local version of the repository, you must first run the docker image without the local repository mounted so that the C extensions can be compiled and then copied over to your local version.
```bash
First run the docker image without mounting externally
docker run --gpus '"device=0"' -it \ -p 8888:8888 \ $USER/deepcell-tf:latest-gpu
Use ctrl-p, ctrl-q (or ctrl+p+q) to exit the running docker image without shutting it down
Then, get the container_id corresponding to the running image of DeepCell-tf
container_id=$(docker ps -q --filter ancestor="$USER/deepcell-tf")
Copy the compiled c extensions into your local version of the codebase:
docker cp "$containerid:/usr/local/lib/python3.6/dist-packages/deepcell/utils/computeoverlap.cpython-36m-x8664-linux-gnu.so" deepcell/utils/computeoverlap.cpython-36m-x86_64-linux-gnu.so
close the running docker
docker kill $container_id
you can now start the docker image with the code mounted for easy editing
docker run --gpus '"device=0"' -it \ -p 8888:8888 \ -v $PWD/deepcell:/usr/local/lib/python3.6/dist-packages/deepcell/ \ -v $PWD/notebooks:/notebooks \ -v $PWD:/data \ $USER/deepcell-tf:latest-gpu ```
How to Cite
- Deep Learning Automates the Quantitative Analysis of Individual Cells in Live-Cell Imaging Experiments
- Accurate cell tracking and lineage construction in live-cell imaging experiments with deep learning
- DeepCell Kiosk: scaling deep learning–enabled cellular image analysis with Kubernetes
- Whole-cell segmentation of tissue images with human-level performance using large-scale data annotation and deep learning
Copyright
Copyright © 2016-2024 The Van Valen Lab at the California Institute of Technology (Caltech), with support from the Shurl and Kay Curci Foundation, Google Research Cloud, the Paul Allen Family Foundation, & National Institutes of Health (NIH) under Grant U24CA224309-01. All rights reserved.
License
This software is licensed under a modified APACHE2. See LICENSE for full details.
Trademarks
All other trademarks referenced herein are the property of their respective owners.
Credits
Owner
- Name: vanvalenlab
- Login: vanvalenlab
- Kind: organization
- Website: https://vanvalen.caltech.edu/
- Repositories: 35
- Profile: https://github.com/vanvalenlab
GitHub Events
Total
- Issues event: 11
- Watch event: 40
- Issue comment event: 10
- Pull request event: 1
- Fork event: 8
Last Year
- Issues event: 11
- Watch event: 40
- Issue comment event: 10
- Pull request event: 1
- Fork event: 8
Committers
Last synced: 9 months ago
Top Committers
| Name | Commits | |
|---|---|---|
| willgraf | 7****f | 260 |
| Morgan Schwartz | m****1@g****m | 40 |
| Ross Barnowski | r****r@c****u | 34 |
| Noah Greenwald | n****d@g****m | 27 |
| MekWarrior | m****k@g****m | 13 |
| vanvalen | v****n@g****m | 12 |
| Dylan Bannon | d****5@g****m | 6 |
| William Graf | W****f@k****g | 4 |
| nitzanrazin | 4****n | 3 |
| Emily Laubscher | 4****h | 2 |
| Jing-Ding | 3****g | 2 |
| Sunny Cui | c****i@c****u | 2 |
| cdpavelchek | 5****k | 2 |
| cpavelchek | c****h@u****u | 2 |
| Josh Moore | j****e@d****k | 1 |
| Kevin Huang | k****8@g****m | 1 |
| Pavel T | p****v@g****m | 1 |
| Shivam Patel | p****9@g****m | 1 |
| Uriah Israel | u****l@g****m | 1 |
| dependabot[bot] | 4****] | 1 |
| jizhouh | 4****h | 1 |
| Kevin Huang | k****g@c****u | 1 |
| rdilip | 9****p | 1 |
Committer Domains (Top 20 + Academic)
Issues and Pull Requests
Last synced: 6 months ago
All Time
- Total issues: 98
- Total pull requests: 79
- Average time to close issues: 17 days
- Average time to close pull requests: 15 days
- Total issue authors: 65
- Total pull request authors: 13
- Average comments per issue: 2.88
- Average comments per pull request: 0.54
- Merged pull requests: 70
- Bot issues: 0
- Bot pull requests: 0
Past Year
- Issues: 8
- Pull requests: 2
- Average time to close issues: 5 days
- Average time to close pull requests: N/A
- Issue authors: 8
- Pull request authors: 1
- Average comments per issue: 1.25
- Average comments per pull request: 0.0
- Merged pull requests: 0
- Bot issues: 0
- Bot pull requests: 0
Top Authors
Issue Authors
- rossbar (9)
- jgu13 (5)
- msschwartz21 (5)
- FiReTiTi (4)
- AlexInJar (4)
- ashishshel (3)
- tbudiman7 (2)
- megleila (2)
- vanvalen (2)
- E5ten (2)
- wzr0108 (2)
- jayluo2 (2)
- acycliq (2)
- adrianomartinelli (2)
- wivan993 (1)
Pull Request Authors
- rossbar (41)
- msschwartz21 (26)
- vanvalen (4)
- elaubsch (3)
- ngreenwald (2)
- willgraf (2)
- kuang-da (2)
- raffeyC (2)
- MekWarrior (1)
- curlup (1)
- ulisrael (1)
- joshmoore (1)
- rdilip (1)
- 251925209391 (1)
Top Labels
Issue Labels
Pull Request Labels
Packages
- Total packages: 3
-
Total downloads:
- pypi 757 last-month
-
Total dependent packages: 3
(may contain duplicates) -
Total dependent repositories: 8
(may contain duplicates) - Total versions: 69
- Total maintainers: 2
pypi.org: deepcell
Deep learning for single cell image segmentation
- Homepage: https://github.com/vanvalenlab/deepcell-tf
- Documentation: https://deepcell.readthedocs.io/
- License: LICENSE
-
Latest release: 0.12.10
published over 1 year ago
Rankings
Maintainers (1)
pypi.org: deepcelltl
Deep learning for single cell image segmentation
- Homepage: https://github.com/vanvalenlab/deepcell-tf
- Documentation: https://deepcelltl.readthedocs.io/
- License: LICENSE
-
Latest release: 0.12.5
published about 3 years ago
Rankings
Maintainers (1)
pypi.org: deepcell-cpu
Deep learning for single cell image segmentation
- Homepage: https://github.com/vanvalenlab/deepcell-tf
- Documentation: https://deepcell-cpu.readthedocs.io/
- License: LICENSE
-
Latest release: 0.12.10
published over 1 year ago
Rankings
Maintainers (1)
Dependencies
- actions/checkout v2 composite
- docker/build-push-action v2 composite
- docker/login-action v1 composite
- docker/setup-buildx-action v1 composite
- docker/setup-qemu-action v1 composite
- actions/checkout v3 composite
- actions/setup-python v4 composite
- actions/checkout v2 composite
- actions/setup-python v2 composite
- pypa/gh-action-pypi-publish master composite
- release-drafter/release-drafter v5 composite
- actions/checkout v2 composite
- actions/setup-python v2 composite
- actions/cache v2 composite
- actions/checkout v2 composite
- actions/setup-python v2 composite
- coverallsapp/github-action master composite
- docker/setup-buildx-action v1 composite
- docker/setup-qemu-action v1 composite
- tensorflow/tensorflow ${TF_VERSION} build
- Sphinx <6
- docutils *
- ipython *
- m2r2 *
- mock *
- nbsphinx *
- nbsphinx-link *
- sphinx-rtd-theme *
- coveralls * test
- pytest * test
- pytest-cov * test
- pytest-mock * test
- ruff * test
- deepcell-toolbox *
- deepcell-tracking *
- jupyterlab *
- matplotlib *
- numpy >=1.16.6
- pydot >=1.4.2,<2
- scikit-image >=0.14.5
- scikit-learn >=0.20.4
- scipy >=1.2.3,<2
- spektral *
- tensorflow *
- tensorflow_addons *
- deepcell-toolbox *
- deepcell-tracking *
- jupyter >=1.0.0,<2
- matplotlib *
- numpy >=1.16.6
- opencv-python-headless <5
- pydot >=1.4.2,<2
- scikit-image >=0.14.5
- scikit-learn >=0.20.4
- scipy >=1.2.3,<2
- spektral *
- tensorflow *
- tensorflow_addons *



