dnachisel
:pencil2: A versatile DNA sequence optimizer
Science Score: 36.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
○CITATION.cff file
-
✓codemeta.json file
Found codemeta.json file -
✓.zenodo.json file
Found .zenodo.json file -
○DOI references
-
○Academic publication links
-
✓Committers with academic emails
1 of 13 committers (7.7%) from academic institutions -
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (14.0%) to scientific vocabulary
Keywords
bioinformatics
codon-optimization
dna-optimization
sequence-design
synbio
synthetic-biology
Last synced: 6 months ago
·
JSON representation
Repository
:pencil2: A versatile DNA sequence optimizer
Basic Info
- Host: GitHub
- Owner: Edinburgh-Genome-Foundry
- License: mit
- Language: Python
- Default Branch: master
- Homepage: https://edinburgh-genome-foundry.github.io/DnaChisel/
- Size: 9.29 MB
Statistics
- Stars: 246
- Watchers: 8
- Forks: 49
- Open Issues: 16
- Releases: 15
Topics
bioinformatics
codon-optimization
dna-optimization
sequence-design
synbio
synthetic-biology
Created over 8 years ago
· Last pushed 10 months ago
Metadata Files
Readme
Changelog
License
README.rst
.. raw:: html

DNA Chisel - a versatile sequence optimizer
===========================================
.. image:: https://github.com/Edinburgh-Genome-Foundry/DnaChisel/actions/workflows/build.yml/badge.svg
:target: https://github.com/Edinburgh-Genome-Foundry/DnaChisel/actions/workflows/build.yml
:alt: GitHub CI build status
.. image:: https://coveralls.io/repos/github/Edinburgh-Genome-Foundry/DnaChisel/badge.svg?branch=master
:target: https://coveralls.io/github/Edinburgh-Genome-Foundry/DnaChisel?branch=master
DNA Chisel (complete documentation `here `_.
The library comes with over 15 classes of sequence specifications which can be
composed to, for instance, codon-optimize genes, meet the constraints of a
commercial DNA provider, avoid homologies between sequences, tune GC content,
or all of this at once! Users can also define their own specifications using
Python, making the library suitable for a large range of automated sequence
design applications, and complex custom design projects. A specification can be
either a hard constraint, which must be satisfied in the final sequence, or an
optimization objective, whose score must be maximized.
For more information, please see the publication.
Citation
--------
DNA Chisel, a versatile sequence optimizer, *Valentin Zulkower, Susan Rosser.* `Bioinformatics `_ (2020) 36, 16, 4508–4509
Usage
-----
Defining a problem via scripts
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
The example below will generate a random sequence and optimize it so that:
- It will be rid of BsaI sites (on both strands).
- GC content will be between 30% and 70% on every 50bp window.
- The reading frame at position 500-1400 will be codon-optimized for *E. coli*.
.. code:: python
from dnachisel import *
# DEFINE THE OPTIMIZATION PROBLEM
problem = DnaOptimizationProblem(
sequence=random_dna_sequence(10000),
constraints=[
AvoidPattern("BsaI_site"),
EnforceGCContent(mini=0.3, maxi=0.7, window=50),
EnforceTranslation(location=(500, 1400))
],
objectives=[CodonOptimize(species='e_coli', location=(500, 1400))]
) # Note: always use a codon optimisation specification with EnforceTranslation
# SOLVE THE CONSTRAINTS, OPTIMIZE WITH RESPECT TO THE OBJECTIVE
problem.resolve_constraints()
problem.optimize()
# PRINT SUMMARIES TO CHECK THAT CONSTRAINTS PASS
print(problem.constraints_text_summary())
print(problem.objectives_text_summary())
# GET THE FINAL SEQUENCE (AS STRING OR ANNOTATED BIOPYTHON RECORDS)
final_sequence = problem.sequence # string
final_record = problem.to_record(with_sequence_edits=True)
Defining a problem via Genbank features
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
You can also define a problem by annotating directly a Genbank as follows:
.. raw:: html
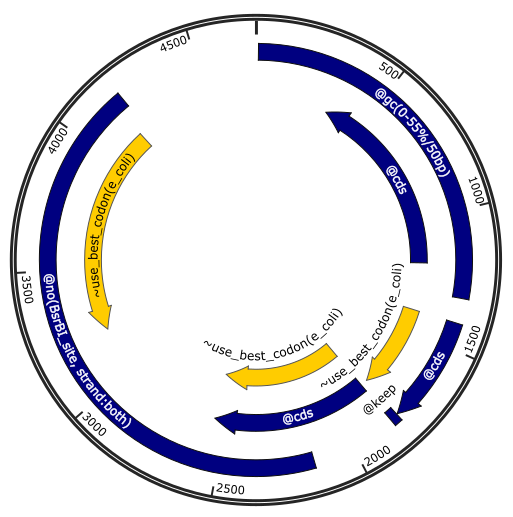
Note that constraints (colored in blue in the illustration) are features of type
``misc_feature`` with a prefix ``@`` followed by the name of the constraints
and its parameters, which are the same as in python scripts. Optimization
objectives (colored in yellow in the illustration) use prefix ``~``. See
`the Genbank API documentation `_
for more details.
Genbank files with specification annotations can be directly fed to the
`web application `_
or processed via the command line interface:
.. code:: bash
# Output the result to "optimized_record.gb"
dnachisel annotated_record.gb optimized_record.gb
Or via a Python script:
.. code:: python
from dnachisel import DnaOptimizationProblem
problem = DnaOptimizationProblem.from_record("my_record.gb")
problem.optimize_with_report(target="report.zip")
By default, only the built-in specifications of DNA Chisel can be used in the
annotations, however it is easy to add your own specifications to the Genbank
parser, and build applications supporting custom specifications on top of
DNA Chisel.
Reports
~~~~~~~
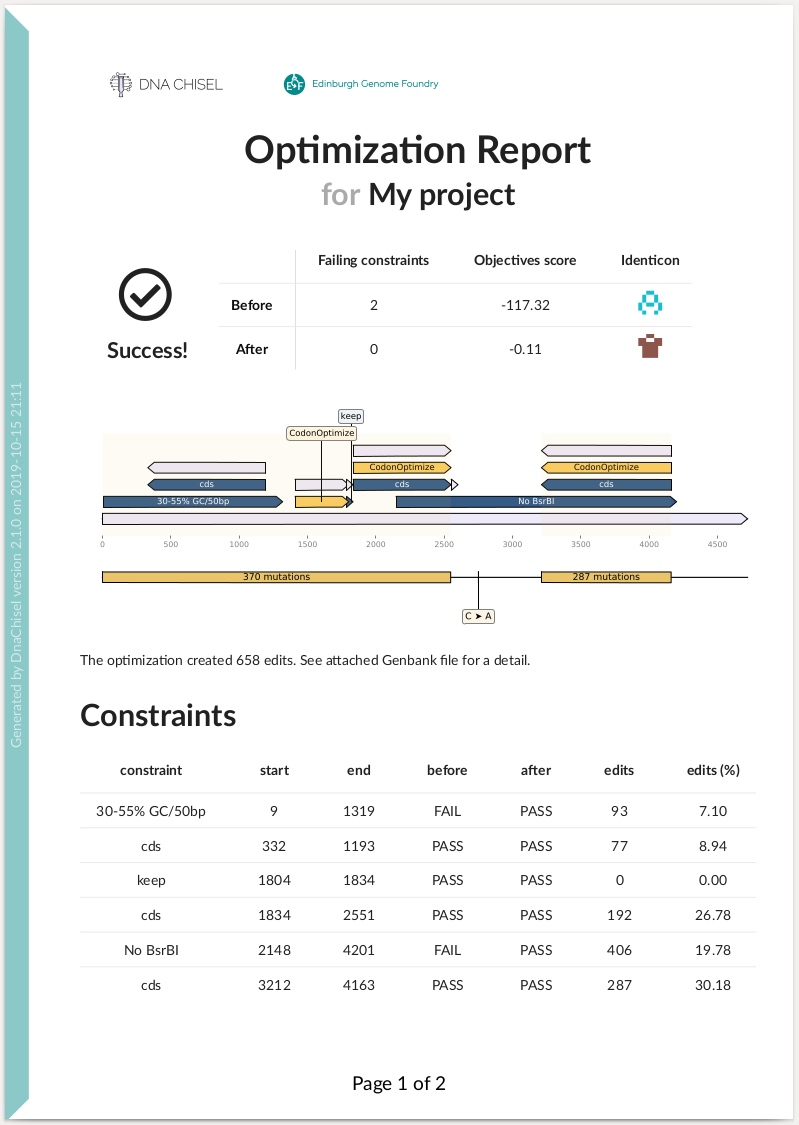
DNA Chisel also implements features for verification and troubleshooting. For
instance by generating optimization reports:
.. code:: python
problem = DnaOptimizationProblem(...)
problem.optimize_with_report(target="report.zip")
Here is an example of summary report:
.. raw:: html
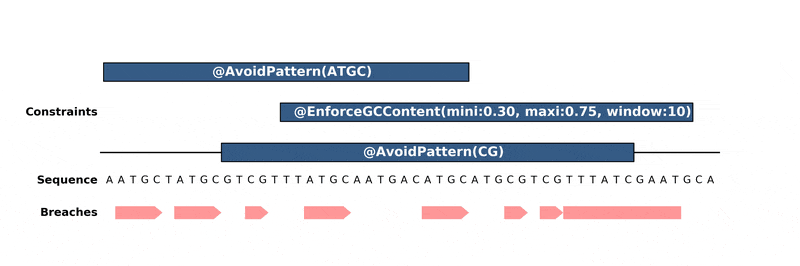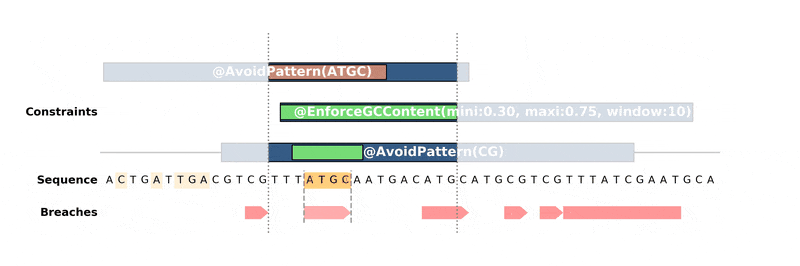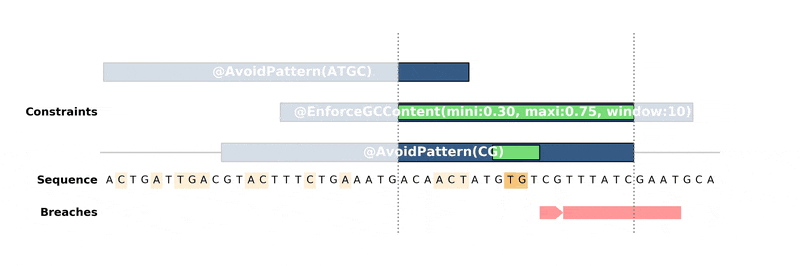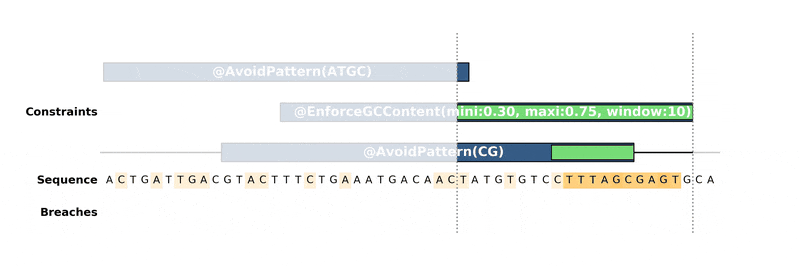
How it works
------------
DNA Chisel hunts down every constraint breach and suboptimal region by
recreating local version of the problem around these regions. Each type of
constraint can be locally *reduced* and solved in its own way, to ensure fast
and reliable resolution.
Below is an animation of the algorithm in action:
.. raw:: html
Installation
------------
DNA Chisel requires Python 3, and can be installed via a pip command:
.. code::
pip install dnachisel # <= minimal install without reports support
pip install 'dnachisel[reports]' # <= full install with all dependencies
The full installation using ``dnachisel[reports]`` downloads heavier libraries
(Matplotlib, PDF reports, sequenticon) for report generation, but is highly
recommended to use DNA Chisel interactively via Python scripts. Also install
`GeneBlocks `_ and its
dependencies if you wish to include a plot of sequence edits in the report.
Optionally, also install Bowtie to be able to use ``AvoidMatches`` (which
removes short homologies with existing genomes). On Ubuntu:
.. code::
sudo apt-get install bowtie
License = MIT
-------------
DNA Chisel is an open-source software originally written at the `Edinburgh Genome Foundry
`_ by `Zulko `_
and `released on Github `_ under the MIT licence (Copyright 2017 Edinburgh Genome Foundry, University of Edinburgh). Everyone is welcome to contribute!
More biology software
---------------------
.. image:: https://raw.githubusercontent.com/Edinburgh-Genome-Foundry/Edinburgh-Genome-Foundry.github.io/master/static/imgs/logos/egf-codon-horizontal.png
:target: https://edinburgh-genome-foundry.github.io/
DNA Chisel is part of the `EGF Codons `_.
- `dnachisel-dtailor-mode `_ brings
features from `D-tailor `_
to DNA Chisel, in particular for the generation of large collection of sequences
covering the objectives fitness landscape (i.e. with sequences with are good at
some objectives and bad at others, and vice versa).
Owner
- Name: Edinburgh Genome Foundry
- Login: Edinburgh-Genome-Foundry
- Kind: organization
- Email: egf-software@ed.ac.uk
- Location: Edinburgh, UK
- Website: https://edinburgh-genome-foundry.github.io/
- Twitter: edingenfoundry
- Repositories: 69
- Profile: https://github.com/Edinburgh-Genome-Foundry
GitHub Events
Total
- Issues event: 12
- Watch event: 25
- Issue comment event: 7
- Push event: 2
- Pull request event: 3
- Fork event: 9
- Create event: 1
Last Year
- Issues event: 12
- Watch event: 25
- Issue comment event: 7
- Push event: 2
- Pull request event: 3
- Fork event: 9
- Create event: 1
Committers
Last synced: 11 months ago
Top Committers
| Name | Commits | |
|---|---|---|
| Zulko | v****r@g****m | 295 |
| Peter Vegh | p****h@p****k | 70 |
| Josh Soref | j****f | 27 |
| Li Xing | l****1@g****m | 8 |
| Brett Hannigan | b****n@g****m | 8 |
| Maoz Gelbart | 1****t | 6 |
| Laura Luebbert | 5****t | 4 |
| Valentin Zulkower | v****r@g****m | 3 |
| Ondrej Sladky | o****y@e****m | 2 |
| Ubuntu | u****u@i****l | 2 |
| Simone Pignotti | s****i@e****m | 1 |
| Max Campbell | 4****1 | 1 |
| Sukolsak Sakshuwong | f****k@p****g | 1 |
Committer Domains (Top 20 + Academic)
Issues and Pull Requests
Last synced: 6 months ago
All Time
- Total issues: 65
- Total pull requests: 36
- Average time to close issues: 2 months
- Average time to close pull requests: 8 days
- Total issue authors: 39
- Total pull request authors: 14
- Average comments per issue: 4.37
- Average comments per pull request: 1.97
- Merged pull requests: 25
- Bot issues: 0
- Bot pull requests: 0
Past Year
- Issues: 12
- Pull requests: 5
- Average time to close issues: 2 months
- Average time to close pull requests: about 23 hours
- Issue authors: 9
- Pull request authors: 3
- Average comments per issue: 0.58
- Average comments per pull request: 0.4
- Merged pull requests: 4
- Bot issues: 0
- Bot pull requests: 0
Top Authors
Issue Authors
- Lix1993 (6)
- y9c (5)
- simone-pignotti (3)
- veghp (3)
- lebolo (3)
- GC-repeat (2)
- ghost (2)
- wyattxuanyang (2)
- eggrandio (2)
- kmcgrathgenerate (2)
- andrewshvv (2)
- lifefoundry-scott (2)
- jlerman44 (2)
- ewallace (2)
- deto (2)
Pull Request Authors
- Lix1993 (13)
- MaozGelbart (6)
- Zulko (4)
- ondrej-sladky-eligo (4)
- veghp (4)
- godotgildor (2)
- simone-pignotti (2)
- maxall41 (2)
- tdsmith (1)
- sukolsak (1)
- chillwei (1)
- rfuisz (1)
- jsoref (1)
Top Labels
Issue Labels
bug (3)
enhancement (2)
Pull Request Labels
Packages
- Total packages: 1
-
Total downloads:
- pypi 50,766 last-month
- Total dependent packages: 6
- Total dependent repositories: 6
- Total versions: 45
- Total maintainers: 1
pypi.org: dnachisel
Optimize DNA sequences under constraints.
- Homepage: https://github.com/Edinburgh-Genome-Foundry/dnachisel
- Documentation: https://dnachisel.readthedocs.io/
- License: mit
-
Latest release: 3.2.16
published 10 months ago
Rankings
Dependent packages count: 1.9%
Downloads: 3.5%
Average: 4.6%
Stargazers count: 5.1%
Dependent repos count: 6.0%
Forks count: 6.8%
Maintainers (1)
Last synced:
6 months ago
Dependencies
setup.py
pypi
- Biopython *
- docopt *
- flametree *
- numpy *
- proglog *
- python_codon_tables *