Science Score: 10.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
○CITATION.cff file
-
○codemeta.json file
-
○.zenodo.json file
-
○DOI references
-
○Academic publication links
-
✓Committers with academic emails
1 of 5 committers (20.0%) from academic institutions -
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (18.2%) to scientific vocabulary
Keywords
bioinformatics
bioinformatics-tool
dna-processing
feature-engineering
feature-extraction
genomics
hashing
hashing-algorithms
k-mer
k-mer-counting
kmer
kmer-counting
kmer-frequency-count
kmers
ngram
ngrams
protein-sequences
rcpp
rcppparallel
rpackage
Last synced: 6 months ago
·
JSON representation
Repository
fast and comprehensive k-mer counting package
Basic Info
- Host: GitHub
- Owner: slowikj
- Language: C++
- Default Branch: master
- Homepage: https://slowikj.github.io/seqR/
- Size: 1.72 MB
Statistics
- Stars: 18
- Watchers: 2
- Forks: 1
- Open Issues: 7
- Releases: 0
Topics
bioinformatics
bioinformatics-tool
dna-processing
feature-engineering
feature-extraction
genomics
hashing
hashing-algorithms
k-mer
k-mer-counting
kmer
kmer-counting
kmer-frequency-count
kmers
ngram
ngrams
protein-sequences
rcpp
rcppparallel
rpackage
Created over 6 years ago
· Last pushed over 4 years ago
Metadata Files
Readme
README.Rmd
---
output: github_document
---
```{r setup, include=FALSE}
knitr::opts_chunk$set(
echo = TRUE,
collapse = TRUE,
comment = "#>",
out.width = "100%"
)
```
```{r, include = FALSE}
library(seqR)
```
# seqR - fast and comprehensive k-mer counting package
[](https://cran.r-project.org/package=seqR)
[](https://github.com/slowikj/seqR/actions)
[](https://lifecycle.r-lib.org/articles/stages.html#stable)
[](https://www.gnu.org/licenses/gpl-3.0)
[](https://codecov.io/github/slowikj/seqR?branch=master)
[](https://www.code-inspector.com/project/23909/status/svg)
[](https://www.code-inspector.com/project/23909/score/svg)
## About
`seqR` is an R package for fast k-mer counting. It provides
* **highly optimized** (the core algorithm is written in C++)
* **in-memory**
* **probabilistic** (with configurable dimensionality of a hash value
used for storing k-mers internally),
* **multi-threaded** (with a configurable size of the batch of sequences (`batch_size`) to process in a single step. If `batch_size` equals 1, the multi-threaded mode is disabled, which potentially causes a longer computation time)
implementation that supports
* **various variants of k-mers** (contiguous, gapped, and positional counterparts)
* **all biological sequences** (e.g., nucleic acids and proteins)
Moreover, the result optimizes memory consumption by the application of **sparse matrices**
(see [package Matrix](https://CRAN.R-project.org/package=Matrix)),
compatible with machine learning packages
such as [ranger](https://CRAN.R-project.org/package=ranger)
and [xgboost](https://CRAN.R-project.org/package=xgboost).
## How to...
### How to install
To install `seqR` from CRAN:
```{r, eval=FALSE}
install.packages("seqR")
```
Alternatively, if you want to use the latest development version:
```{r, eval=FALSE}
# install.packages("devtools")
devtools::install_github("slowikj/seqR")
```
### How to use
The package provides two functions that facilitate k-mer counting
* `count_kmers` (used for counting k-mers of one type)
* `count_multimers` (a wrapper of `count_kmers`, used for counting k-mers of many types in a single invocation of the function)
and one function used for custom processing of k-mer matrices:
* `rbind_columnwise` (a helper function used for merging several k-mer matrices that do not have same sets of columns)
To learn more, see [features overview vignette](https://slowikj.github.io/seqR/articles/features-overview.html)
and [reference](https://slowikj.github.io/seqR/reference/index.html).
#### Examples
##### counting 5-mers
```{r}
count_kmers(sequences=c("AAAAAVVAVFF", "DFGSADFGSA"),
k=5)
```
##### counting gapped 5-mers with gaps (0, 1, 0, 2) (XX_XX__X)
```{r}
count_kmers(sequences=c("AAAAAVVAVFF", "DFGSADFGSA"),
kmer_gaps=c(0, 1, 0, 2))
```
##### counting 1-mers and 2-mers
```{r}
data(CsgA)
CsgA[1L:2]
count_multimers(sequences=CsgA,
k_vector = c(1, 2))
```
### How to cite
For citation type:
```{r, eval=FALSE}
citation("seqR")
```
or use:
Jadwiga Słowik and Michał Burdukiewicz (2021). seqR: fast and comprehensive k-mer counting package. R package version 1.0.0.
## Benchmarks
The `seqR` package has been compared with other existing k-mer counting R packages:
[biogram](https://CRAN.R-project.org/package=biogram),
[kmer](https://CRAN.R-project.org/package=kmer),
[seqinr](https://CRAN.R-project.org/package=seqinr),
and [biostrings](https://bioconductor.org/packages/Biostrings).
All benchmark experiments have been performed using Intel Core i7-6700HQ 2.60GHz 8 cores, using the [microbenchmark](https://CRAN.R-project.org/package=microbenchmark) R package.
### Contiguous k-mers
#### Changing k
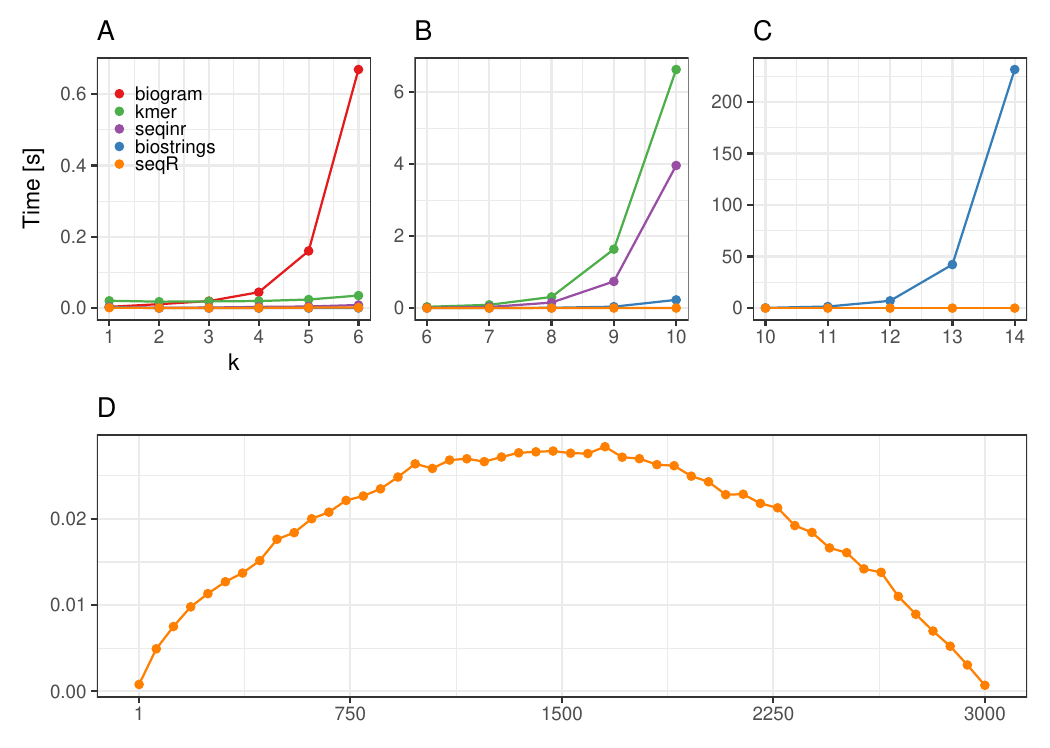 The input consists of one `DNA` sequence of length `3 000`.
#### Changing the number of sequences
The input consists of one `DNA` sequence of length `3 000`.
#### Changing the number of sequences
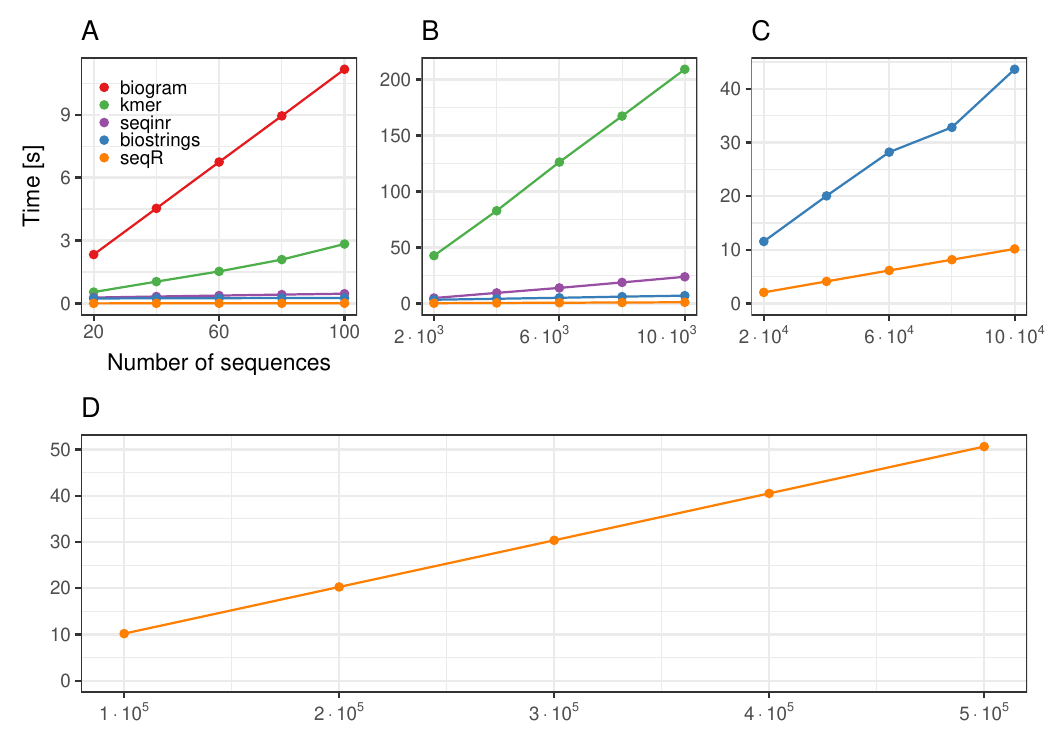 Each `DNA` sequence has `3 000` elements, `contiguous 5-mer` counting.
### Gapped k-mers
#### Changing the first contiguous part of a k-mer
Each `DNA` sequence has `3 000` elements, `contiguous 5-mer` counting.
### Gapped k-mers
#### Changing the first contiguous part of a k-mer
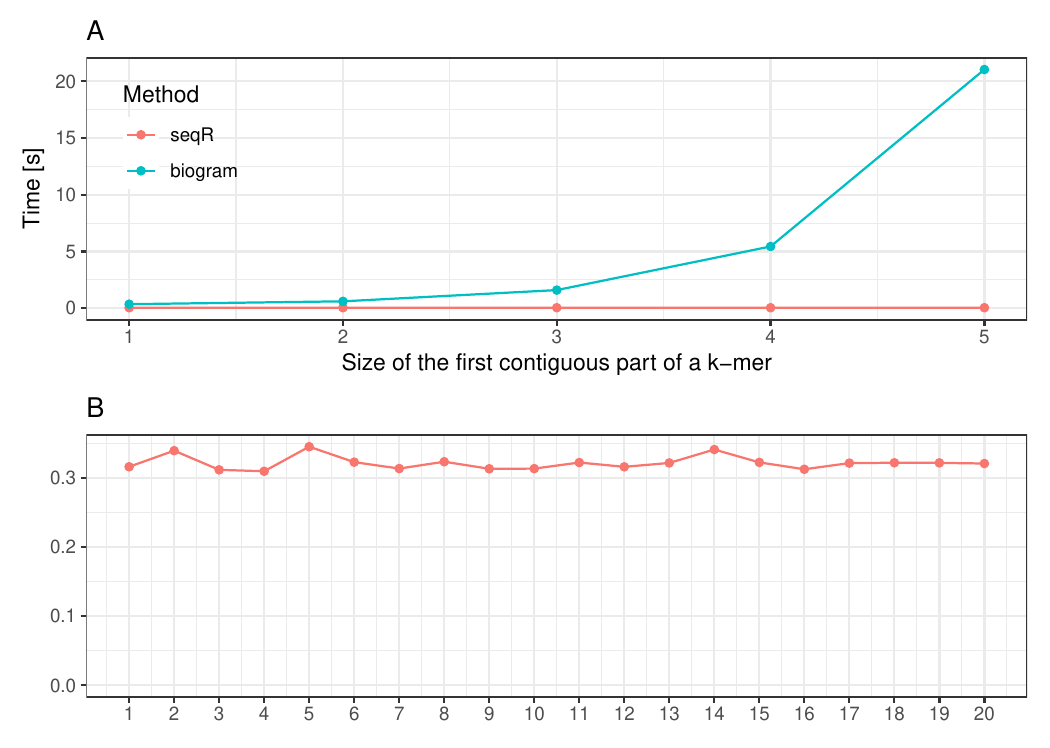 The input consists of one `DNA` sequence of length `1 000 000`. `Gapped 5-mers` counting with base gaps `(1, 0, 0, 1)`.
#### Changing the first gap size
The input consists of one `DNA` sequence of length `1 000 000`. `Gapped 5-mers` counting with base gaps `(1, 0, 0, 1)`.
#### Changing the first gap size
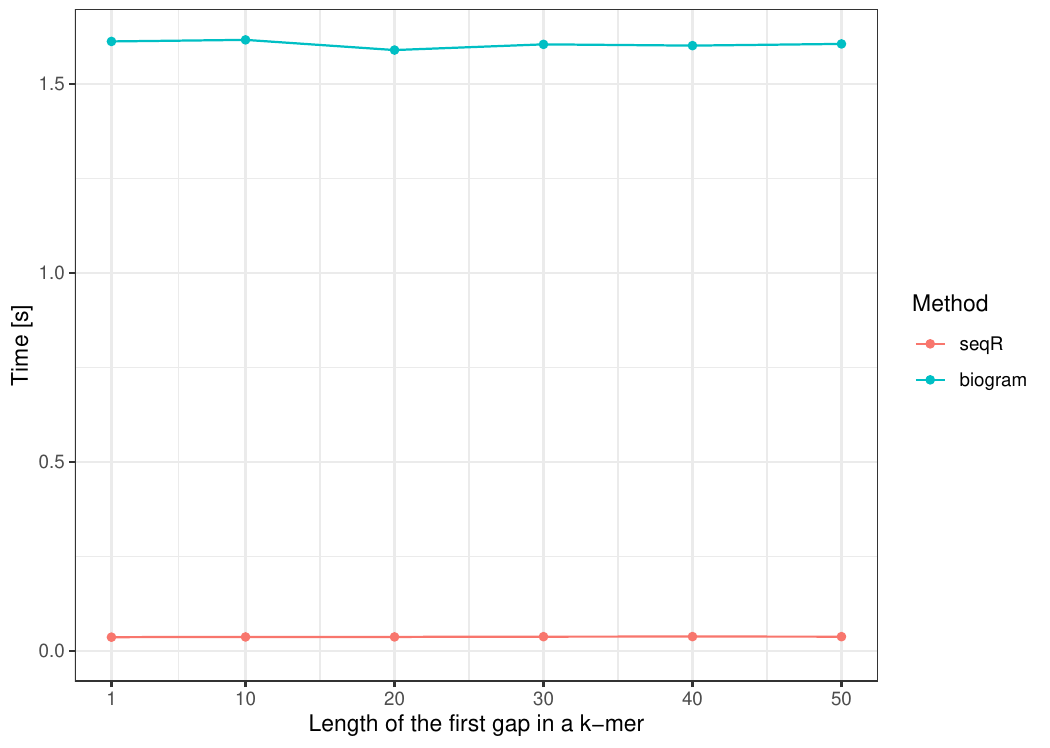 The input consists of one `DNA` sequence of length `100 000`. `Gapped 5-mers` counting with base gaps `(1, 0, 0, 1)`.
The input consists of one `DNA` sequence of length `100 000`. `Gapped 5-mers` counting with base gaps `(1, 0, 0, 1)`.
Owner
- Name: Jadwiga Słowik
- Login: slowikj
- Kind: user
- Location: Poland
- Website: https://onlinejudge.org/index.php?option=onlinejudge&page=show_authorstats&userid=57739
- Twitter: slowikj5
- Repositories: 2
- Profile: https://github.com/slowikj
passionate software developer & data scientist, problem solver, competitive programmer
GitHub Events
Total
- Watch event: 1
Last Year
- Watch event: 1
Committers
Last synced: over 2 years ago
Top Committers
| Name | Commits | |
|---|---|---|
| Jadwiga Słowik | j****k@g****m | 383 |
| Jadwiga Słowik | j****5@g****m | 54 |
| slowikj | s****j@s****l | 10 |
| Michał Burdukiewicz | m****z@g****m | 7 |
| piotr-ole | p****k@g****m | 3 |
Committer Domains (Top 20 + Academic)
Issues and Pull Requests
Last synced: over 1 year ago
All Time
- Total issues: 49
- Total pull requests: 43
- Average time to close issues: 3 months
- Average time to close pull requests: 2 days
- Total issue authors: 2
- Total pull request authors: 1
- Average comments per issue: 0.29
- Average comments per pull request: 0.0
- Merged pull requests: 40
- Bot issues: 0
- Bot pull requests: 0
Past Year
- Issues: 0
- Pull requests: 0
- Average time to close issues: N/A
- Average time to close pull requests: N/A
- Issue authors: 0
- Pull request authors: 0
- Average comments per issue: 0
- Average comments per pull request: 0
- Merged pull requests: 0
- Bot issues: 0
- Bot pull requests: 0
Top Authors
Issue Authors
- slowikj (46)
- michbur (1)
Pull Request Authors
- slowikj (43)
Top Labels
Issue Labels
enhancement (23)
performance (11)
bug (8)
refactor (5)
hashing (4)
wontfix (3)
documentation (2)
nice to have (2)
Pull Request Labels
Packages
- Total packages: 1
- Total downloads: unknown
- Total dependent packages: 0
- Total dependent repositories: 1
- Total versions: 2
cran.r-project.org: seqR
Fast and Comprehensive K-Mer Counting Package
- Homepage: https://github.com/slowikj/seqR
- Documentation: http://cran.r-project.org/web/packages/seqR/seqR.pdf
- License: GPL (≥ 3)
- Status: removed
-
Latest release: 1.0.1
published over 4 years ago
Rankings
Stargazers count: 16.3%
Forks count: 21.9%
Dependent repos count: 25.5%
Dependent packages count: 29.8%
Average: 36.6%
Downloads: 89.7%
Last synced:
11 months ago
Dependencies
DESCRIPTION
cran
- R >= 3.6 depends
- Matrix * imports
- Rcpp >= 1.0.4 imports
- RcppParallel >= 5.1.2 imports
- rlang * imports
- slam * imports
- covr * suggests
- ggplot2 * suggests
- knitr * suggests
- microbenchmark * suggests
- pryr * suggests
- rmarkdown * suggests
- spelling * suggests
- testthat * suggests