deepbedmap
Going beyond BEDMAP2 using a super resolution deep neural network. Also a convenient flat file data repository for high resolution bed elevation datasets around Antarctica.
Science Score: 23.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
○CITATION.cff file
-
○codemeta.json file
-
○.zenodo.json file
-
✓DOI references
Found 7 DOI reference(s) in README -
✓Academic publication links
Links to: zenodo.org -
○Committers with academic emails
-
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (11.0%) to scientific vocabulary
Keywords
Repository
Going beyond BEDMAP2 using a super resolution deep neural network. Also a convenient flat file data repository for high resolution bed elevation datasets around Antarctica.
Basic Info
Statistics
- Stars: 43
- Watchers: 5
- Forks: 26
- Open Issues: 6
- Releases: 0
Topics
Metadata Files
README.md
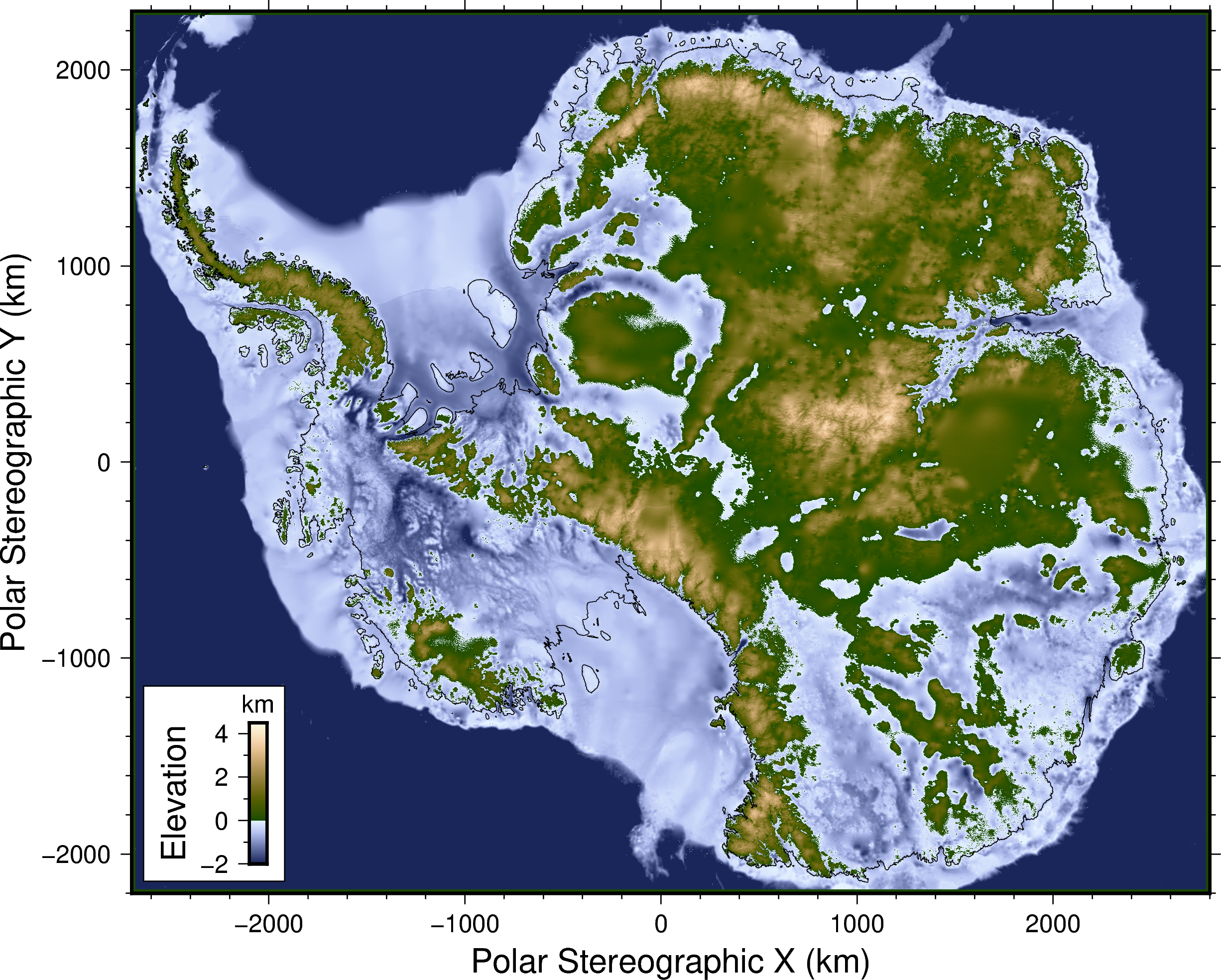
DeepBedMap [paper] [poster] [presentation]
Going beyond BEDMAP2 using a super resolution deep neural network. Also a convenient flat file data repository for high resolution bed elevation datasets around Antarctica.
Directory structure
``` deepbedmap/ ├── features/ (files describing the high level behaviour of various features) │ ├── *.feature... (easily understandable specifications written using the Given-When-Then gherkin language) │ └── README.md (markdown information on the feature files) ├── highres/ (contains high resolution localized DEMs) │ ├── *.txt/csv/grd/xyz... (input vector file containing the point-based bed elevation data) │ ├── *.json (the pipeline file used to process the xyz point data) │ ├── *.nc (output raster netcdf files) │ └── README.md (markdown information on highres data sources) ├── lowres/ (contains low resolution whole-continent DEMs) │ ├── bedmap2_bed.tif (the low resolution DEM!) │ └── README.md (markdown information on lowres data sources) ├── misc/ (miscellaneous raster datasets) │ ├── *.tif (Surface DEMs, Ice Flow Velocity, etc. See list in Issue #9) │ └── README.md (markdown information on miscellaneous data sources) ├── model/ (*hidden in git, neural network model related files) │ ├── train/ (a place to store the raster tile bounds and model training data) │ └── weights/ (contains the neural network model's architecture and weights) ├── .env (environment variable config file used by pipenv) ├── .Getting started
Quickstart
Launch in Binder (Interactive jupyter notebook/lab environment in the cloud).
Installation
Start by cloning this repo-url
git clone <repo-url>
Then I recommend using conda to install the non-python binaries (e.g. GMT, CUDA, etc). The conda virtual environment will also be created with Python and pipenv installed.
cd deepbedmap
conda env create -f environment.yml
Activate the conda environment first.
conda activate deepbedmap
Then set some environment variables before using pipenv to install the necessary python libraries,
otherwise you may encounter some problems (see Common problems below).
You may want to ensure that which pipenv returns something similar to ~/.conda/envs/deepbedmap/bin/pipenv.
export HDF5_DIR=$CONDA_PREFIX/
export LD_LIBRARY_PATH=$CONDA_PREFIX/lib/
pipenv install --python $CONDA_PREFIX/bin/python --dev
#or just
HDF5_DIR=$CONDA_PREFIX/ LD_LIBRARY_PATH=$CONDA_PREFIX/lib/ pipenv install --python $CONDA_PREFIX/bin/python --dev
Finally, double-check that the libraries have been installed.
pipenv graph
Syncing/Updating to new dependencies
conda env update -f environment.yml
pipenv sync --dev
Common problems
Note that the .env file stores some environment variables.
So if you run conda activate deepbedmap followed by some other command and get an ...error while loading shared libraries: libpython3.7m.so.1.0...,
you may need to run pipenv shell or do pipenv run <cmd> to have those environment variables registered properly.
Or just run this first:
export LD_LIBRARY_PATH=$CONDA_PREFIX/lib/
Also, if you get a problem when using pipenv to install netcdf4, make sure you have done:
export HDF5_DIR=$CONDA_PREFIX/
and then you can try using pipenv install or pipenv sync again.
See also this issue for more information.
Running jupyter lab
conda activate deepbedmap
pipenv shell
python -m ipykernel install --user --name deepbedmap #to install conda env properly
jupyter kernelspec list --json #see if kernel is installed
jupyter lab &
Citing
The paper is published at The Cryosphere and can be referred to using the following BibTeX code:
@Article{tc-14-3687-2020,
AUTHOR = {Leong, W. J. and Horgan, H. J.},
TITLE = {DeepBedMap: a deep neural network for resolving the bed topography of Antarctica},
JOURNAL = {The Cryosphere},
VOLUME = {14},
YEAR = {2020},
NUMBER = {11},
PAGES = {3687--3705},
URL = {https://tc.copernicus.org/articles/14/3687/2020/},
DOI = {10.5194/tc-14-3687-2020}
}
The DeepBedMap_DEM v1.1.0 dataset is available from Zenodo at https://doi.org/10.5281/zenodo.4054246. Neural network model training experiment runs are also recorded at https://www.comet.ml/weiji14/deepbedmap. Python code for the DeepBedMap model here on Github is also mirrored on Zenodo at https://doi.org/10.5281/zenodo.3752613.
Owner
- Name: Wei Ji
- Login: weiji14
- Kind: user
- Location: Wellington
- Company: @developmentseed
- Website: https://weiji14.xyz
- Repositories: 67
- Profile: https://github.com/weiji14
Geospatial Data Scientist/ML Practitioner @developmentseed. Towards GPU-native and cloud-native geospatial machine learning!
GitHub Events
Total
- Watch event: 2
Last Year
- Watch event: 2
Committers
Last synced: 7 months ago
Top Committers
| Name | Commits | |
|---|---|---|
| Wei Ji | w****g@v****z | 379 |
| dependabot[bot] | s****t@d****m | 84 |
| dependabot[bot] | d****] | 2 |
| Gunesh Shanbhag | 1****5 | 1 |
| stickler-ci | s****t@s****m | 1 |
Committer Domains (Top 20 + Academic)
Issues and Pull Requests
Last synced: 6 months ago
All Time
- Total issues: 36
- Total pull requests: 158
- Average time to close issues: about 2 months
- Average time to close pull requests: 5 days
- Total issue authors: 5
- Total pull request authors: 4
- Average comments per issue: 0.58
- Average comments per pull request: 0.73
- Merged pull requests: 126
- Bot issues: 20
- Bot pull requests: 113
Past Year
- Issues: 0
- Pull requests: 0
- Average time to close issues: N/A
- Average time to close pull requests: N/A
- Issue authors: 0
- Pull request authors: 0
- Average comments per issue: 0
- Average comments per pull request: 0
- Merged pull requests: 0
- Bot issues: 0
- Bot pull requests: 0
Top Authors
Issue Authors
- dependabot-preview[bot] (20)
- weiji14 (13)
- nififikan (1)
- psychonaute (1)
Pull Request Authors
- dependabot-preview[bot] (112)
- weiji14 (44)
- stickler-ci[bot] (1)
- gshanbhag525 (1)
Top Labels
Issue Labels
Pull Request Labels
Packages
- Total packages: 1
- Total downloads: unknown
- Total dependent packages: 0
- Total dependent repositories: 0
- Total versions: 14
proxy.golang.org: github.com/weiji14/deepbedmap
- Documentation: https://pkg.go.dev/github.com/weiji14/deepbedmap#section-documentation
- License: lgpl-3.0
-
Latest release: v1.1.0
published over 5 years ago
Rankings
Dependencies
- cuda_driver 440.64.*
- cudatoolkit 10.0.*
- gmt 6.0.0.*
- libspatialindex 1.9.0.*
- pip 19.2.1.*
- pipenv 2020.6.2.*
- python 3.7.3.*
- behave ==1.2.6 develop
- nbval ==0.9.2 develop
- pytest ==5.0.1 develop
- black ==19.3b0
- chainer ==7.0.0
- comet-ml ==2.0.5
- cupy-cuda100 ==7.0.0
- dask ==2.1.0
- descartes ==1.1.0
- filelock ==3.2
- geopandas ==0.5.1
- joblib ==0.13.2
- jupyterlab ==1.0.2
- jupytext ==1.2.0
- livelossplot ==0.4.1
- matplotlib ==3.1.1
- netcdf4 ==1.4.1
- numpy ==1.17.3
- optuna ==2.0.0
- pandas ==0.25.0
- pygmt ==0.1.1
- quilt ==2.9.15
- rasterio ==1.0.24
- rtree ==0.8.3
- salem ==0.2.4
- scikit-image ==0.15.0
- ssim-chainer https://github.com/higumachan/ssim-chainer.git#9c54f25b7c068b562b711027a92c346ce0708b52
- tqdm ==4.32.2
- xarray ==0.12.3
- 189 dependencies



