plantFEM
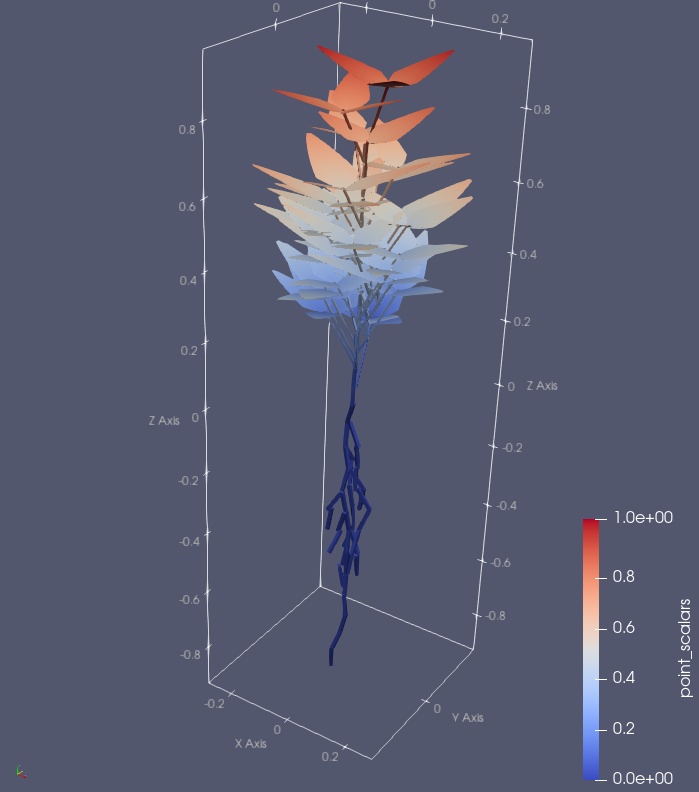
This is a plant/farming simulator based on Finite Element Method, which targets crops in fields and soil foundations. This software provides multi-physical simulations of agriculture for canopies, plants, and organs for farmers, breeders, agronomists, and civil engineers. Please try and give us feedback.
Science Score: 46.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
○CITATION.cff file
-
✓codemeta.json file
Found codemeta.json file -
✓.zenodo.json file
Found .zenodo.json file -
○DOI references
-
✓Academic publication links
Links to: scholar.google -
✓Committers with academic emails
1 of 5 committers (20.0%) from academic institutions -
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (10.4%) to scientific vocabulary
Keywords
Repository
This is a plant/farming simulator based on Finite Element Method, which targets crops in fields and soil foundations. This software provides multi-physical simulations of agriculture for canopies, plants, and organs for farmers, breeders, agronomists, and civil engineers. Please try and give us feedback.
Basic Info
- Host: GitHub
- Owner: kazulagi
- License: mit
- Language: Fortran
- Default Branch: master
- Homepage: https://github.com/kazulagi/plantFEM.git
- Size: 305 MB
Statistics
- Stars: 44
- Watchers: 3
- Forks: 5
- Open Issues: 4
- Releases: 3
Topics
Metadata Files
README.md


 <!--
<!--
 -->
-->
 <!--
<!--
 -->
-->

[New!] plantFEM 22.04(LTS) is released!
Specification
| plantFEM | |
| ---- | ---- |
| Developer | Haruka Tomobe & plantFEM.org |
| Working state | current |
| Written in | Fortran 2003, Python 3.x, C89 |
|Source model | Open-source |
| Initial release | 21.10 (20 October 2021)|
| First Long-Term Surpport (LTS) release | 22.04 (23 April 2022) |
| Repository | https://github.com/kazulagi/plantfem |
| Usage | Agricultural CAE, Digital Twins for Agricultural/Civil Engineering |
| Target | Personal computers, HPC-Clusters, Servers |
| Package Manager | soja (experimental) |
|Platforms | x86-64 |
|Default user interface | CLI |
| Lisence | MIT |
| Community | Slack (private channel) |
| Official Website | plantFEM.org |
| Objects | Simulation | Simulation (experimental) | | ---- | ---- | ---- | | Elementary entities | Pseudo-static Deformation, Diffusion | Contact, Dynamic deformation, Reaction-diffusion | | Soybean | Creation, Measure size, Measure mass | Deformation, Contact, Photosynthesis | | Grape | Creation, Measure size, Measure mass | Deformation, Contact, Photosynthesis | | Maize | Creation, Measure size, Measure mass | Deformation, Contact, Photosynthesis |
| Library structure | |
| ---- | ---- |
| std | Extention of Fortran 2003. Contains fundamental classes for file-IOs and Mathematical operations.|
| fem | Library for implementing Finite Element Method. You can create meshes (FEMDomain), shape-functions, boundary conditions, initial conditions and some elemental matrices.|
| sim | A set of simulators for FEMDomain. Contains deformation, diffusion, and some experimental implementations. |
| obj | A set of classes for realistic agricultural high-/low-level objects. High-level objects: Soil, Soybean, Maize, Grape...etc. Low-level objects: stem, leaf, air, light ...etc. |
| IO formats | | | ---- | ---- | | Input | json, vtk, msh, ASCII-text| | Output | json, vtk, msh, stl, ply, ASCII-text|
| Commands | |
| ---- | ---- |
| plantfem search | Search sample codes by a keyword |
| plantfem install | Build library and setting PATH |
| plantfem build | Build server.f90 and creates executable file server.out |
| plantfem run | = plantfem build && mpirun ./server.out |
| plantfem man | Manual for plantfem command. |
| Finite Elements | | | ---- | ---- | | 2-node line element | 1D 2D 3D| | 4-node isoparametric element | 2D| | 8-node isoparametric element | 3D|
| System Requirements | | | ---- | ---- | | Operation System | Linux (Ubuntu 16.04+, ElementaryOS, LinuxMint, Debian), Windows 10/11 (with WSL-Ubuntu), macOS | | CPU | 2 cores, 1.4 GHz| | RAM | 2 GB | | Storage | 1 GB |
| Dependancies (minimal) | version | | ---- | ---- | | git | 2.25.1 | | Python | 3.4 or later |
| Dependancies (Installed by setup/setup.py) | version, info | | ---- | ---- | | gcc | 6.4.0 or later | | gfortran | 4.8.0 or later (Fortran 2003 or later) | | mpif90 | = OpenMPI compiler, 2.x or later | | apt | 1.2.35 or later | | pip | pip3 or later | | curl | 7.47.0 or later |
What's next plantFEM 22.10 ?
APIs for Python, C/C++, and JavaScript (Experimental).
Documentation
Click here!
For detail, you can create documentation by
ford ford.md
How to install
Clone the repository.
git clone https://github.com/kazulagi/plantFEM.gitRun
python3 install.py. The default compiler ismpif90. If you want to use Intel compiler, runpython3 install.py --compiler=intelinstead of it.
For Windows users:
Activate your WSL2 (Windows 10)
Install "Ubuntu 20.04" from Microsoft Store
Run command
wget https://plantfem.org/download/plantfem_22.04-ubuntu2004_amd64.deb
sudo apt install plantfem_22.04-ubuntu2004_amd64.deb
- You can open files by this command
explorer.exe .
- Enjoy!
For Ubuntu users:
You can download pre-build packages for
[Click to download] Ubuntu 18.04
[Click to download] Ubuntu 20.04
In case you are using Ubuntu 18.04, execute the next one-liner.
wget https://plantfem.org/download/plantfem_22.04-ubuntu1804_amd64.deb && sudo apt install plantfem_22.04-ubuntu1804_amd64.deb
If you are using Ubuntu 20.04, execute the next one-liner.
wget https://plantfem.org/download/plantfem_22.04-ubuntu2004_amd64.deb && sudo apt install plantfem_22.04-ubuntu2004_amd64.deb
If you want to build and run as a docker container,
(1) Activate "Docker for Windows" (https://docs.docker.com/docker-for-windows/)
(2) Open command-prompt and run
{.copy}
git clone -b 22.04 https://github.com/kazulagi/plantFEM && cd plantFEM/docker
How to Use
- Search sample codes
You can search sample codes by
plantfem search
and type your keywords.
Open editors (e.g. VSCode) and edit&save it with extention of
.f90Build your script (For example,
test.f90) by
plantfem load test.f90
plantfem build
- Run your script.
./server.out
Or you can run it with multi-core workstations or HPC-clusters.
- Execute
plantfem init
to initialize directory.
Edit
server.f90Build the project by
plantfem deploy
- Run it by
mpirun --hostfile [your hostfile for OpenMPI] -np [number of process] ./server.out
Here is an example of hostfile
hostfile
192.168.0.1 cpu=6
192.168.0.2 cpu=6
192.168.0.3 cpu=6
call plantfem from Python (experimental)
```python
import plantfem as pf
soy = pf.soybean(name="hellosoy") soy.create(config="./plantfem/Tutorial/obj/realSoybeanConfig.json") soy.msh(name="hellosoy") soy.json(name="hello_soy")
soy.stl(name="hello_soy")
path to plantfem
soy.run(path="./plantfem")
```
Try it now
Plant simulator based on Finite Element Method (FEM).
- Tutorial: Click here!
How to add modules for plantFEM?
(1) Create your Fortran add-on in plantfem/addon or other places. An example is shown in addon/addon_example.f90.
```Fortran module addonexample use plantfem type::addonexample_ ! Member variables real(real64),private :: realVal real(int32 ),private :: intVal contains ! methods (publicname => privatename) procedure :: set => setaddonsample procedure :: show => showaddonsample end type contains
! Definitions of methods
! ################################################ subroutine setaddonsample(obj,realVal, intVal) class(addonexample_),intent(inout) :: obj real(real64),optional,intent(in) :: realVal integer(int32),optional,intent(in) :: intVal
obj%realVal = input(default=0.0d0, option=realVal)
obj%intVal = input(default=0, option=intVal)
end subroutine ! ################################################
! ################################################ subroutine showaddonsample(obj) class(addonexample_),intent(in) :: obj print *, "Real-64bit value is :: ", obj%realVal print *, "int-32bit value is :: ", obj%intVal end subroutine ! ################################################
end module addon_example ```
(2) Compile your addon by typing "addon" after
plantfem
Then, type addon and tap ENTER
```
addon installing add-on Directory path of your awesome addon is : (default path = addon) addon installing from addon addonexample Compiling ./addon/addonexample.f90
addon_example.o | ########################### | (100%)```
(3) Run your script (An example is shown in Tutorial/HowToUseAddon/ex1.f90)
Fortran
program main
use addon_example
implicit none
type(addon_example_) :: obj
call obj%set(realVal=8.0d0, intVal=-100)
call obj%show()
end program
(4) Done!
```
test.f90
Real-64bit value is :: 8.0000000000000000
int-32bit value is :: -100.000000```
You can set a hostfile
vi ./etc/hostfile
and a number of process by
./plantfem cpu-core
or
vi ./etc/cpucore
logs;
- 2019/01/19 :: This Document is written.
- 2019/01/20 :: ControlParameterClass is included
- 2019/01/21 :: Bug Fixed :: FEMDomainClass/ExportFEMDomain.f90 about Neumann-Boundary
- 2019/01/21 :: "Method:DeallocateAll"::Deallocate all alleles.(For all objects)
- 2019/01/25 :: DisplayMesh.f90/ Implement >> Export Mesh data.
- 2019/02/19 :: DiffusionEquationClass.f90 >> Solver for diffusion equations with time-integration by Clank-Nicolson Method
- 2019/03/03 :: FiniteDeformationClass.f90 >> Solver for Finite Deformation problems (2D and 3D).
- 2019/03/10 :: FEMIfaceClass.f90 >> Interface objects (3D).
- 2019/03/10 :: MeshOperationClass.f90 >> Mesh can be devided and interface mesh ca be generated (3D).
- 2019/03/21 :: install.sh and run.sh is created.
- 2019/03/23 :: Standarize FEMDomain(.scf) objects
- 2019/03/24 :: Field class and Simulator Class are created.
- 2019/03/26 :: Interface Solvers are created as MultiPhysics
- 2019/04/13 :: PreprocessingClass is created.
- 2019/04/13 :: DictionaryClass is created.
- 2019/05/13 :: PreProcessingClass is created.
- 2019/06/29 :: ContactMechanicsClass is under debug
- 2019/08/01 :: Jupyter notebook is introduced as GUI.
- 2019/08/01 :: Installer for Windows/macOS/Linux is created.
- 2019/08/03 :: Delauney triangulation is now under development.
- 2019/09/06 :: Bugfix of Simulatior
- 2019/09/23 :: Source code is opened.Now solvers for diffusion, finite deformation, and diffusion-deformation coupling are available. Contact solver is under debugging.
- 2021/05/15 :: Now you can install plantFEM by
install.pyand can run byplantfem run - 2021/10/21 :: Release plantFEM 21.10
- 2022/04/21 :: Beat-release: plantFEM 22.04
Acknowledgements
This project is financially supported by the following research grants.
- Grant-in-Aid for Young Scientists(Start-up), (ID:20K22599), JSPS, JAPAN
- Grant-in-Aid for JSPS Fellows, (ID:17J02383), JSPS, JAPAN
Relevant publications
[1] Haruka Tomobe, Kazunori Fujisawa, Akira Murakami, Experiments and FE-analysis of 2-D root-soil contact problems based on node-to-segment approach, Soils and Foundations, Volume 59, Issue 6, 2019, Pages 1860-1874.
[2] Haruka Tomobe, Kazunori Fujisawa, Akira Murakami, A Mohr-Coulomb-Vilar model for constitutive relationship in root-soil interface under changing suction, Soils and Foundations, Volume 61,2021, Pages 815–835.
[3] Haruka Tomobe, Yu Tanaka, Tomoya Watanabe, plantFEM: A Numerical Platform for Multi-physical Simulation of Plants, Third International Workshop on Machine Learning for Cyber-Agricultural Systems (MLCAS2021), Page 22.
[4] Haruka Tomobe, Vikas Sharma, Harusato Kimura, Hitoshi Morikawa, An Energy-based Overset Finite Element Method for Pseudo-static Structural Analysis. J. Sci. Comput. 2023, 94:55.
Others are under revision and/or under preparation.
TODO:
- Python-interface
- Spline curve
- NURBS curve
Owner
- Name: Haruka Tomobe
- Login: kazulagi
- Kind: user
- Location: Japan
- Company: tomobe.haruka.58m[at]gmail.com
- Website: https://kazulagi.github.io/HarukaTomobe
- Repositories: 34
- Profile: https://github.com/kazulagi
Assistant Professor at Tokyo Institute of Technology, Ph.D. (Agronomy)
GitHub Events
Total
- Watch event: 5
- Delete event: 2
- Issue comment event: 5
- Push event: 70
- Pull request review event: 1
- Pull request event: 67
- Fork event: 1
Last Year
- Watch event: 5
- Delete event: 2
- Issue comment event: 5
- Push event: 70
- Pull request review event: 1
- Pull request event: 67
- Fork event: 1
Committers
Last synced: 7 months ago
Top Committers
| Name | Commits | |
|---|---|---|
| Your Name | y****u@e****m | 340 |
| haruka | t****m@s****p | 323 |
| Haruka Tomobe | t****m@g****m | 199 |
| Stefano Zaghi | s****i@g****m | 33 |
| haruka | h****a@o****l | 1 |
Committer Domains (Top 20 + Academic)
Issues and Pull Requests
Last synced: 6 months ago
All Time
- Total issues: 9
- Total pull requests: 620
- Average time to close issues: over 1 year
- Average time to close pull requests: about 23 hours
- Total issue authors: 3
- Total pull request authors: 3
- Average comments per issue: 0.33
- Average comments per pull request: 0.03
- Merged pull requests: 604
- Bot issues: 0
- Bot pull requests: 5
Past Year
- Issues: 0
- Pull requests: 67
- Average time to close issues: N/A
- Average time to close pull requests: about 6 hours
- Issue authors: 0
- Pull request authors: 1
- Average comments per issue: 0
- Average comments per pull request: 0.1
- Merged pull requests: 58
- Bot issues: 0
- Bot pull requests: 0
Top Authors
Issue Authors
- kazulagi (7)
- Beliavsky (1)
- HowcanoeWang (1)
Pull Request Authors
- kazulagi (614)
- dependabot[bot] (5)
- r1wtn (1)
Top Labels
Issue Labels
Pull Request Labels
Packages
- Total packages: 2
- Total downloads: unknown
-
Total dependent packages: 0
(may contain duplicates) -
Total dependent repositories: 0
(may contain duplicates) - Total versions: 2
proxy.golang.org: github.com/kazulagi/plantfem
- Documentation: https://pkg.go.dev/github.com/kazulagi/plantfem#section-documentation
- License: mit
-
Latest release: v0.0.1-alpha
published over 6 years ago
Rankings
proxy.golang.org: github.com/kazulagi/plantFEM
- Documentation: https://pkg.go.dev/github.com/kazulagi/plantFEM#section-documentation
- License: mit
-
Latest release: v0.0.1-alpha
published over 6 years ago
Rankings
Dependencies
- ubuntu 18.04 build
- ubuntu 18.04 build
- jaraco.packaging >=8.2 test
- pytest >=4.6 test
- pytest-black >=0.3.7 test
- pytest-checkdocs >=2.4 test
- pytest-cov * test
- pytest-enabler >=1.0.1 test
- pytest-flake8 * test
- pytest-mypy * test
- pytest-virtualenv * test
- rst.linker >=1.9 test
- sphinx * test
- fastapi ==0.65.2
- pydantic ==1.7.4
- pytest ==6.2.2
- uvicorn ==0.13
- jaraco.packaging >=8.2 test
- pytest >=4.6 test
- pytest-black >=0.3.7 test
- pytest-checkdocs >=2.4 test
- pytest-cov * test
- pytest-enabler >=1.0.1 test
- pytest-flake8 * test
- pytest-mypy * test
- pytest-virtualenv * test
- rst.linker >=1.9 test
- sphinx * test
- fastapi ==0.65.2
- pydantic ==1.7.4
- pytest ==6.2.2
- uvicorn ==0.13