divbrowse
A web application for interactive visualization and exploratory data analysis of variant call matrices
Science Score: 57.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
✓CITATION.cff file
Found CITATION.cff file -
✓codemeta.json file
Found codemeta.json file -
✓.zenodo.json file
Found .zenodo.json file -
✓DOI references
Found 4 DOI reference(s) in README -
○Academic publication links
-
○Committers with academic emails
-
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (15.0%) to scientific vocabulary
Keywords
Repository
A web application for interactive visualization and exploratory data analysis of variant call matrices
Basic Info
- Host: GitHub
- Owner: IPK-BIT
- License: mit
- Language: Svelte
- Default Branch: main
- Homepage: https://divbrowse.ipk-gatersleben.de
- Size: 2.6 MB
Statistics
- Stars: 20
- Watchers: 2
- Forks: 3
- Open Issues: 5
- Releases: 2
Topics
Metadata Files
README.md

Website: https://divbrowse.ipk-gatersleben.de
Documentation: https://divbrowse.readthedocs.io
Paper: https://doi.org/10.1093/gigascience/giad025
Table of contents: - About DivBrowse - Installation - Try out DivBrowse - Screenshots - Usage workflow concept - Architecture
About DivBrowse
DivBrowse is a web application for interactive exploration and analysis of very large SNP matrices.
It offers a novel approach for interactive visualization and analysis of genomic diversity data and optionally also gene annotation data. The use of standard file formats for data input supports interoperability and seamless deployment of application instances based on established bioinformatics pipelines. The possible integration into 3rd-party web applications supports interoperability and reusability.
The integrated ad-hoc calculation of variant summary statistics and principal component analysis enables the user to perform interactive analysis of population structure for single genetic features like genes, exons and promoter regions. Data interoperability is achieved by the possibility to export genomic diversity data for genomic regions of interest in standardized VCF files.
Installation
The installation via pip or container images is described in the documentation: https://divbrowse.readthedocs.io/en/stable/installation.html
Try out DivBrowse
If you want to test DivBrowse please visit the demo instances listed here: https://divbrowse.ipk-gatersleben.de/#demo-instances
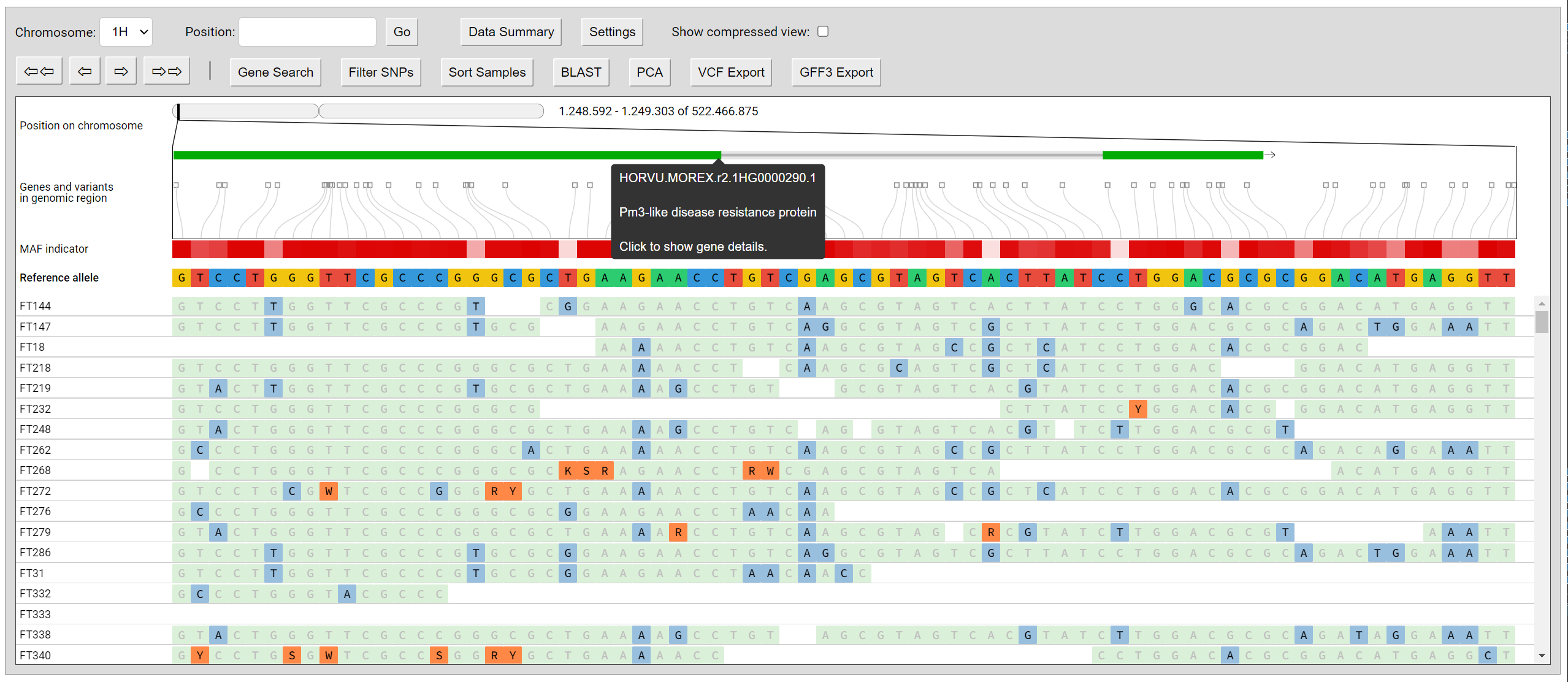
Screenshots
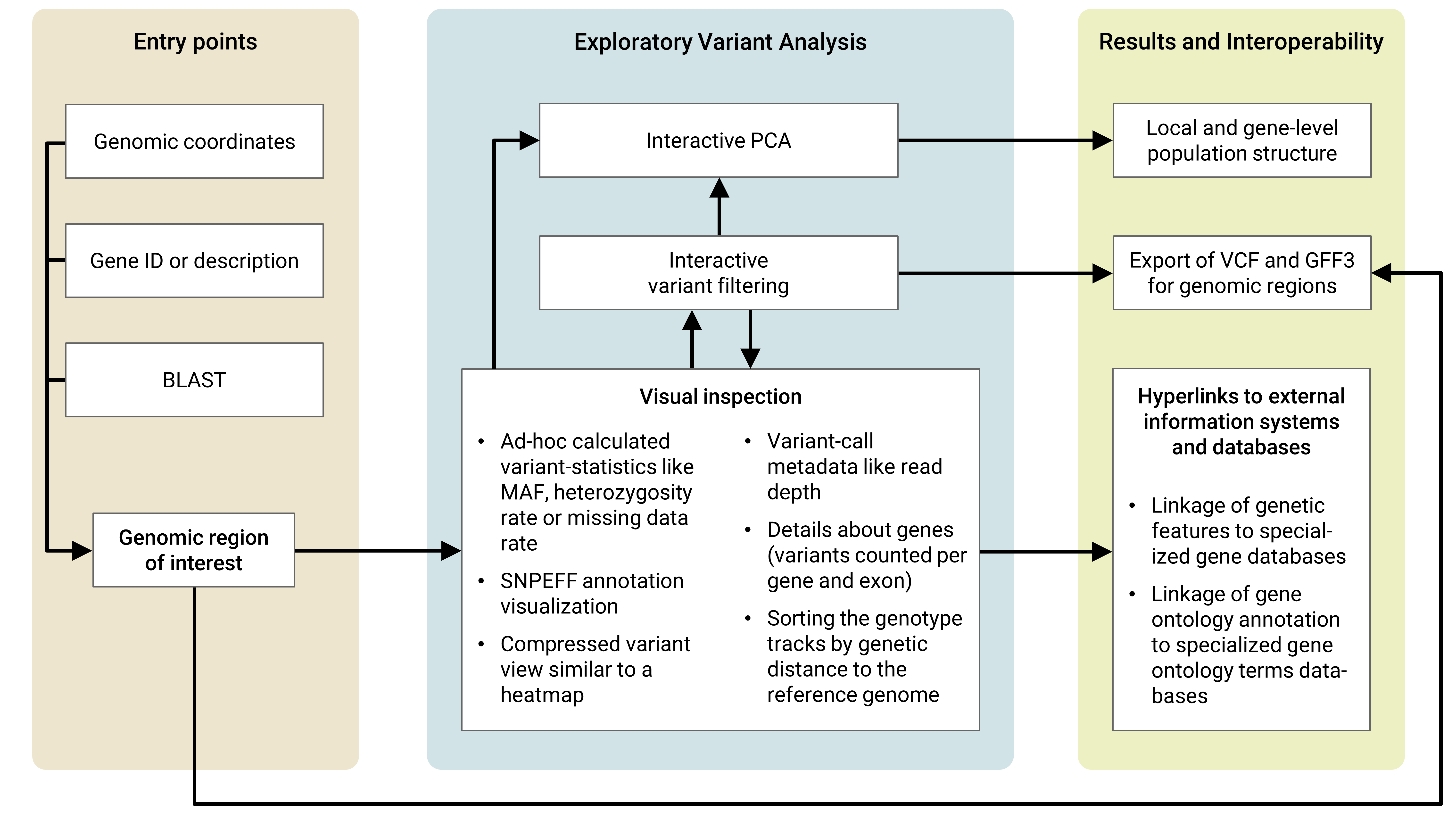
Usage workflow concept
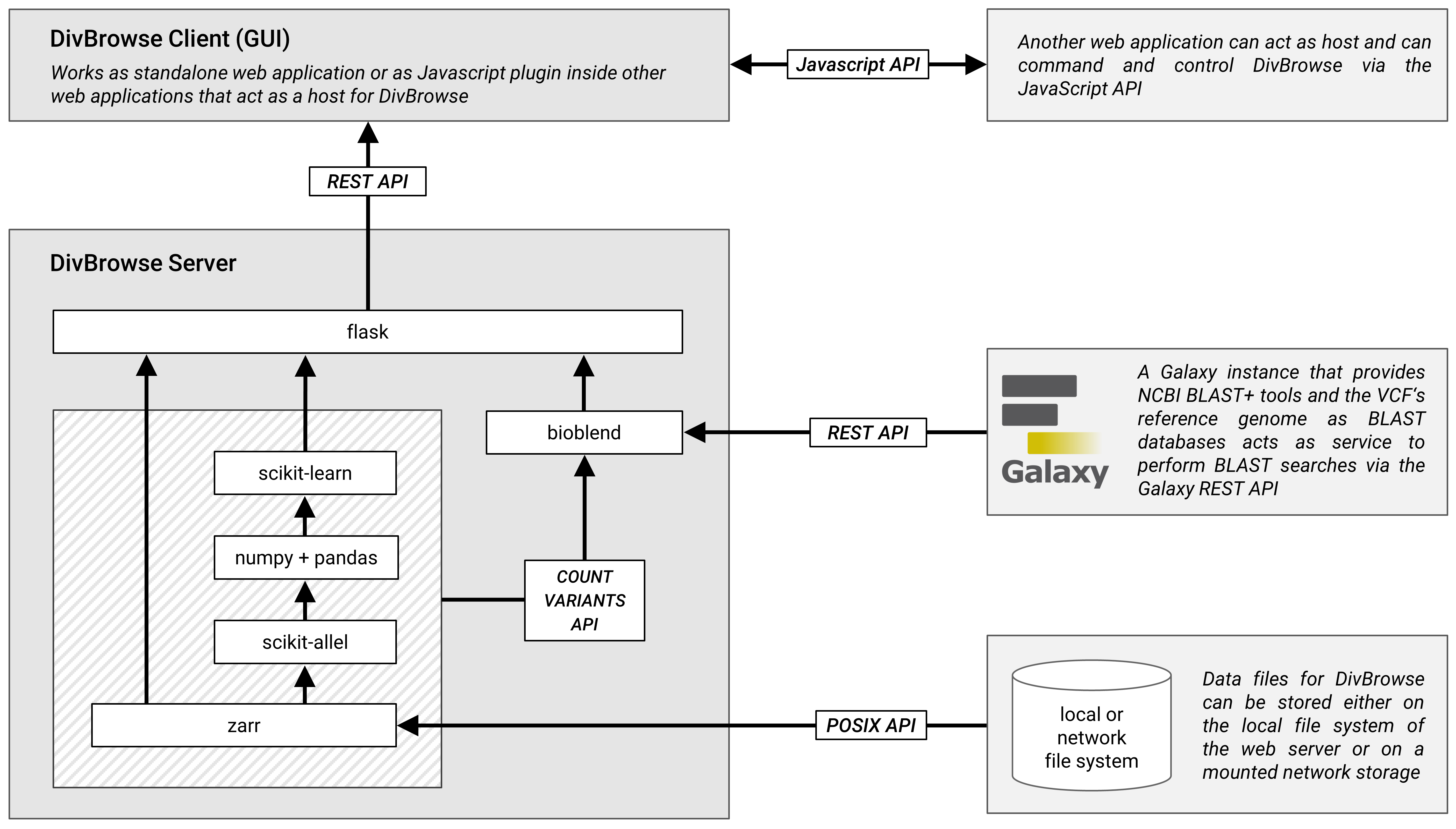
Architecture
Owner
- Name: IPK Gatersleben - Bioinformatics and Information Technology
- Login: IPK-BIT
- Kind: organization
- Location: Germany
- Website: https://www.ipk-gatersleben.de/en/breeding-research/bioinformatics-and-information-technology/
- Repositories: 3
- Profile: https://github.com/IPK-BIT
Leibniz Institute of Plant Genetics and Crop Plant Research (IPK) Gatersleben
Citation (CITATION.cff)
# This CITATION.cff file was generated with cffinit.
# Visit https://bit.ly/cffinit to generate yours today!
cff-version: 1.2.0
title: >-
DivBrowse - interactive visualization and
exploratory data analysis of variant call matrices
message: >-
If you use this software, please cite it using the
metadata from this file.
preferred-citation:
authors:
- given-names: Patrick
family-names: König
email: koenig@ipk-gatersleben.de
orcid: 'https://orcid.org/0000-0002-8948-6793'
affiliation: >-
Leibniz Institute of Plant Genetics and Crop
Plant Research (IPK) Gatersleben, 06466
Seeland, Germany
- given-names: Sebastian
family-names: Beier
orcid: 'https://orcid.org/0000-0002-2177-8781'
affiliation: >-
Leibniz Institute of Plant Genetics and Crop
Plant Research (IPK) Gatersleben, 06466
Seeland, Germany
email: s.beier@fz-juelich.de
- given-names: Martin
family-names: Mascher
email: mascher@ipk-gatersleben.de
affiliation: >-
Leibniz Institute of Plant Genetics and Crop
Plant Research (IPK) Gatersleben, 06466
Seeland, Germany
orcid: 'https://orcid.org/0000-0001-6373-6013'
- given-names: Nils
family-names: Stein
orcid: 'https://orcid.org/0000-0003-3011-8731'
affiliation: >-
Leibniz Institute of Plant Genetics and Crop
Plant Research (IPK) Gatersleben, 06466
Seeland, Germany
email: stein@ipk-gatersleben.de
- given-names: Matthias
family-names: Lange
email: lange@ipk-gatersleben.de
orcid: 'https://orcid.org/0000-0002-4316-078X'
affiliation: >-
Leibniz Institute of Plant Genetics and Crop
Plant Research (IPK) Gatersleben, 06466
Seeland, Germany
- given-names: Uwe
family-names: Scholz
orcid: 'https://orcid.org/0000-0001-6113-3518'
email: scholz@ipk-gatersleben.de
affiliation: >-
Leibniz Institute of Plant Genetics and Crop
Plant Research (IPK) Gatersleben, 06466
Seeland, Germany
title: DivBrowse - interactive visualization and exploratory data analysis of variant call matrices
type: article
doi: 10.1093/gigascience/giad025
journal: GigaScience
volume: 12
year: 2023
type: software
authors:
- given-names: Patrick
family-names: König
email: koenig@ipk-gatersleben.de
orcid: 'https://orcid.org/0000-0002-8948-6793'
affiliation: >-
Leibniz Institute of Plant Genetics and Crop
Plant Research (IPK) Gatersleben, 06466
Seeland, Germany
- given-names: Sebastian
family-names: Beier
orcid: 'https://orcid.org/0000-0002-2177-8781'
affiliation: >-
Leibniz Institute of Plant Genetics and Crop
Plant Research (IPK) Gatersleben, 06466
Seeland, Germany
email: s.beier@fz-juelich.de
- given-names: Martin
family-names: Mascher
email: mascher@ipk-gatersleben.de
affiliation: >-
Leibniz Institute of Plant Genetics and Crop
Plant Research (IPK) Gatersleben, 06466
Seeland, Germany
orcid: 'https://orcid.org/0000-0001-6373-6013'
- given-names: Nils
family-names: Stein
orcid: 'https://orcid.org/0000-0003-3011-8731'
affiliation: >-
Leibniz Institute of Plant Genetics and Crop
Plant Research (IPK) Gatersleben, 06466
Seeland, Germany
email: stein@ipk-gatersleben.de
- given-names: Matthias
family-names: Lange
email: lange@ipk-gatersleben.de
orcid: 'https://orcid.org/0000-0002-4316-078X'
affiliation: >-
Leibniz Institute of Plant Genetics and Crop
Plant Research (IPK) Gatersleben, 06466
Seeland, Germany
- given-names: Uwe
family-names: Scholz
orcid: 'https://orcid.org/0000-0001-6113-3518'
email: scholz@ipk-gatersleben.de
affiliation: >-
Leibniz Institute of Plant Genetics and Crop
Plant Research (IPK) Gatersleben, 06466
Seeland, Germany
identifiers:
- type: doi
value: 10.1093/gigascience/giad025
description: The DivBrowse paper on Oxford Academic GigaScience Journal.
- type: url
value: 'https://divbrowse.ipk-gatersleben.de'
description: Project homepage
repository-code: 'https://github.com/IPK-BIT/divbrowse'
url: 'https://divbrowse.ipk-gatersleben.de/'
repository-artifact: 'https://pypi.org/project/divbrowse/'
doi: 10.1093/gigascience/giad025
journal: GigaScience
volume: 12
year: 2023
abstract: >-
DivBrowse is a software for interactive
visualization and analysis of the diversity of
genomic variants. It uses VCF and GFF3 files as
data input and consists of a web server written in
Python and a GUI written in Javascript.
keywords:
- genomics
- data visualization
- variation data
- variant call format
- biodiversity
license: MIT
#version: 1.0.1
date-released: '2023-04-21'
GitHub Events
Total
- Watch event: 4
Last Year
- Watch event: 4
Issues and Pull Requests
Last synced: 6 months ago
All Time
- Total issues: 9
- Total pull requests: 0
- Average time to close issues: 5 months
- Average time to close pull requests: N/A
- Total issue authors: 4
- Total pull request authors: 0
- Average comments per issue: 0.22
- Average comments per pull request: 0
- Merged pull requests: 0
- Bot issues: 0
- Bot pull requests: 0
Past Year
- Issues: 1
- Pull requests: 0
- Average time to close issues: N/A
- Average time to close pull requests: N/A
- Issue authors: 1
- Pull request authors: 0
- Average comments per issue: 0.0
- Average comments per pull request: 0
- Merged pull requests: 0
- Bot issues: 0
- Bot pull requests: 0
Top Authors
Issue Authors
- patrick-koenig (6)
- lukasmueller (1)
- juancresc (1)
- langeipk (1)
Pull Request Authors
Top Labels
Issue Labels
Pull Request Labels
Packages
- Total packages: 1
-
Total downloads:
- pypi 25 last-month
- Total dependent packages: 0
- Total dependent repositories: 1
- Total versions: 8
- Total maintainers: 2
pypi.org: divbrowse
A web application for interactive visualization and analysis of genotypic variant matrices
- Homepage: https://divbrowse.ipk-gatersleben.de/
- Documentation: https://divbrowse.readthedocs.io/
- License: MIT
-
Latest release: 1.1.0
published almost 3 years ago
Rankings
Maintainers (2)
Dependencies
- @rollup/plugin-commonjs ^17.0.0 development
- @rollup/plugin-node-resolve ^11.0.0 development
- @rollup/plugin-replace ^2.3.3 development
- less ^3.11.1 development
- postcss ^8.3.5 development
- rollup ^2.3.4 development
- rollup-plugin-css-only ^3.1.0 development
- rollup-plugin-livereload ^2.0.0 development
- rollup-plugin-root-import ^1.0.0 development
- rollup-plugin-svelte ^7.0.0 development
- rollup-plugin-terser ^7.0.0 development
- svelte ^3.33.0 development
- svelte-preprocess ^4.7.4 development
- @sveltejs/svelte-virtual-list ^3.0.1
- axios ^0.21.1
- d3 ^5.16.0
- dataframe-js ^1.4.4
- fuse.js ^6.4.6
- lodash ^4.17.15
- plotly.js-dist ^1.54.7
- sirv-cli ^1.0.0
- svelte-inview ^1.0.0
- svelte-moveable ^0.8.11
- svelte-range-slider-pips ^1.7.0
- svelte-simple-datatables ^0.1.25
- svelte-simple-modal ^0.6.0
- sveltejs-tippy ^3.0.0
- tippy.js ^6.2.3
- flask *
- sphinx ^4.0.2 develop
- sphinx-autoapi ^1.6.0 develop
- sphinx-click ^3.0.1 develop
- sphinx_rtd_theme ^0.5.2 develop
- bioblend ^0.16.0
- click ^8.0.1
- flask ^2.0.1
- numpy ^1.21.1
- pandas ^1.3.0
- python ^3.9
- pyyaml ^5.4.1
- scikit-allel ^1.3.5
- scikit-learn ^0.24.2
- simplejson ^3.17.3
- sphinx ^4.0.2
- sphinx-autoapi ^1.6.0
- sphinx-click ^3.0.1
- sphinx_rtd_theme ^0.5.2
- tables ^3.6.1
- umap-learn ^0.5.2
- waitress 2.1.2
- zarr ^2.8.3
- actions/checkout master composite
- actions/setup-python v2 composite
- pypa/gh-action-pypi-publish master composite







