https://github.com/leseixas/blendpy
Computational toolkit for investigating thermodynamic models of alloys using first-principles calculations
Science Score: 26.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
○CITATION.cff file
-
✓codemeta.json file
Found codemeta.json file -
○.zenodo.json file
-
✓DOI references
Found 4 DOI reference(s) in README -
○Academic publication links
-
○Academic email domains
-
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (13.7%) to scientific vocabulary
Keywords
Repository
Computational toolkit for investigating thermodynamic models of alloys using first-principles calculations
Basic Info
Statistics
- Stars: 2
- Watchers: 1
- Forks: 0
- Open Issues: 0
- Releases: 55
Topics
Metadata Files
README.md
Blendpy is a computational toolkit for investigating thermodynamic models of alloys using first-principles calculations
Table of contents
Installation
Install blendpy effortlessly using pip, Python’s default package manager, by running the following command in your terminal:
sh
pip install --upgrade pip
pip install blendpy
Usage
This comprehensive tutorial guides you through calculating alloy properties. In this section, you'll learn how to determine key parameters—such as the enthalpy of mixing, and the spinodal and binodal decomposition curves derived from phase diagrams. We start by defining the alloy components, move through geometry optimization, and conclude with advanced modeling techniques using the DSI model.
To start, provide a list of structure files (e.g., CIF or POSCAR) that represent your alloy components. For best accuracy, it is recommended that these files have been pre-optimized using the same calculator and parameters that will be used in the subsequent alloy property calculations.
If you already have these optimized structures, you may skip ahead to the "DSI model" section. If not, proceed to the "Geometry Optimization" section to prepare your structures for analysis.
Geometry optimization
For example, let's calculate the properties of an Au-Pt alloy. We begin by retrieving the Au (fcc) and Pt (fcc) geometries from ASE. Next, we optimize these geometries using the MACE calculator, which leverages machine learning interatomic potentials.[^fn1] Finally, we save the optimized structures for use in the DSI model. To achieve this, we will follow several key steps.
Step 1: Import the necessary modules from ASE and MACE:
python
from ase.io import write
from ase.build import bulk
from ase.optimize import BFGSLineSearch
from ase.filters import UnitCellFilter
from mace.calculators import mace_mp
Step 2: Create Atoms objects for gold (Au) and platinum (Pt) using the bulk function:
```python
Create Au and Pt Atoms object
gold = bulk("Au", cubic=True) platinum = bulk("Pt", cubic=True) ```
Step 3: Create a MACE calculator object to optimize the structures and assign the calculator to the Atoms objects:
```python
calcmace = macemp(model="small",
dispersion=False,
default_dtype="float32",
device='cpu')
Assign the calculator to the Atoms objects
gold.calc = calcmace platinum.calc = calcmace ```
Step 4: Optimize the unit cells of Au and Pt using the BFGSLineSearch optimizer:
```python
Optimize Au and Pt unit cells
optimizergold = BFGSLineSearch(UnitCellFilter(gold)) optimizergold.run(fmax=0.01)
optimizerplatinum = BFGSLineSearch(UnitCellFilter(platinum)) optimizerplatinum.run(fmax=0.01) ```
Step 5: Save the optimized unit cells to CIF files: ```python
Save the optimized unit cells for Au and Pt
write("Aurelaxed.cif", gold) write("Ptrelaxed.cif", platinum) ```
Dilute solution interpolation (DSI) model
Enthalpy of mixing
Import the DSIModel from blendpy and create a DSIModel object using the optimized structures:
```python
from blendpy import DSIModel
Create a DSIModel object
dsimodel = DSIModel(alloycomponents = ['Aurelaxed.cif', 'Ptrelaxed.cif'], supercell = [2,2,2], calculator = calc_mace) ```
Optimize the structures within the DSIModel object:
```python
Optimize the structures
dsi_model.optimize(method=BFGSLineSearch, fmax=0.01, logfile=None) ```
Calculate the enthalpy of mixing for the AuPt alloy: ```python
Calculate the enthalpy of mixing
enthalpyofmixing = dsimodel.getenthalpyofmixing(npoints=101) x = np.linspace(0, 1, len(enthalpyofmixing)) dfenthalpy = pd.DataFrame({'x': x, 'enthalpy': enthalpyof_mixing}) ```
Plotting the enthalpy of mixing ```python import numpy as np import matplotlib.pyplot as plt
fig, ax = plt.subplots(1,1, figsize=(5,5))
ax.setxlabel("$x$", fontsize=20) ax.setylabel("$\Delta H{mix}$ (kJ/mol)", fontsize=20) ax.setxlim(0,1) ax.setylim(-7,7) ax.setxticks(np.linspace(0,1,6)) ax.set_yticks(np.arange(-6,7,2))
Plot the data
color1='#fc4e2a' ax.plot(dfenthalpy['x'][::5], dfenthalpy['enthalpy'][::5], marker='o', color=color1, markersize=8, linewidth=3, zorder=2, label="DSI Model (MACE)")
REFERENCE: Okamoto, H. and Massalski, T., The Au-Pt (gold-platinum) system, Bull. Alloy Phase Diagr. 6, 46-56 (1985).
dfexp = pd.readcsv("data/experimental/expAuPt.csv") ax.plot(dfexp['x'], df_exp['enthalpy'], 's', color='grey', markersize=8, label="Exp. Data", zorder=1) ax.legend(loc="best", fontsize=16)
ax.tickparams(direction='in', axis='both', which='major', labelsize=20, width=3, length=8) ax.setboxaspect(1) for spine in ax.spines.values(): spine.setlinewidth(3)
plt.tight_layout()
plt.savefig("enthalpyofmixing.png", dpi=600, format='png', bbox_inches='tight')
plt.show() ```

Figure 1 - Enthalpy of mixing of the Au-Pt alloy computed using the DSI model and MACE interatomic potentials.
DSI model from pre-calculated data
Using blendpy, we can also calculate the enthalpy of mixing for an alloy based on DFT simulations that are not initiated by the DSIModel class object. Instead, we can use external data for the total energies of the pristine and dilute supercell systems. For instance, using the ab initio simulation software GPAW,[^fn2] we calculate the total energies for the Au-Pt alloy using [3,3,3] supercells of Au and Pt (Atoms(Au27) and Atoms(Pt27)), as well as the dilute systems (Atoms(Au26Pt) and Atoms(Pt26Au)). These total energies are then used to construct the energy matrix (energy_matrix) in the following form:
```python
energy_matrix = np.array([[ -85.940400, -89.230299], # [[ Au27, Au26Pt], [-170.278459, -173.891172]]) # [ Pt26Au, Pt27]] ```
Next, we demonstrate how to calculate the enthalpy of mixing using the input energy matrix.
```python import numpy as np import pandas as pd import matplotlib.pyplot as plt from ase.build import bulk from ase.io import write from blendpy import DSIModel
Create the two bulk structures
gold = bulk('Au', 'fcc') palladium = bulk('Pt', 'fcc')
Write the structures to CIF files
write("ciffiles/Au.cif", gold) write("ciffiles.Pt.cif", palladium)
Create the DSI model
alloycomponents = ['ciffiles/Au.cif', 'ciffiles/Pt.cif'] x0 = 1./27 dsimodel= DSIModel(alloycomponents=alloycomponents, supercell=[3,3,3], x0=x0)
Set the energy matrix
energy_matrix = np.array([[ -85.940400, -89.230299], [-170.278459, -173.891172]])
dsimodel.setenergymatrix(energymatrix)
enthalpy= dsimodel.getenthalpyofmixing()
x = np.linspace(0, 1, len(enthalpy))
df_enthalpy = pd.DataFrame({'x': x, 'enthalpy': enthalpy}) ```
In this case, it is MANDATORY to specify the minimum dilution factor (x0), the supercell size ([3,3,3]) used in the ab initio simulations, and the unit cell files ('Au.cif' and 'Pt.cif'). The unit cell files do not need to match exactly those used in the ab initio simulations.
Finally, we can plot the enthalpy of mixing and compare it with the experimental data.[^fn3] ```python
Plot the enthalpy of mixing
fig, ax = plt.subplots(1,1, figsize=(6,6))
ax.setxlabel('x', fontsize=20) ax.setylabel('$\Delta H{mix}$ (kJ/mol)', fontsize=20) ax.setxlim(0,1) ax.setylim(-7,7) ax.setxticks(np.linspace(0,1,6)) ax.setyticks(np.arange(-6,7,2)) ax.settitle("Au${1-x}$Pt${x}$", fontsize=20, pad=10)
plot data
Experimental
dfexp = pd.readcsv("../../data/experimental/expAuPt.csv") ax.plot(dfexp['x'], df_exp['enthalpy'], 's', color='grey', linestyle='--', linewidth=3, markersize=8, label="Exp. Data", zorder=1)
DSI model
ax.plot(dfenthalpy['x'][::5], dfenthalpy['enthalpy'][::5], marker='o', color='#fc4e2a', markersize=8, linewidth=3, zorder=2, label="DSI model from DFT")
ax.legend(fontsize=16, loc='best') ax.tickparams(axis='both', which='major', labelsize=20, width=3, length=8, direction='in') ax.setbox_aspect(1)
for spine in ax.spines.values(): spine.set_linewidth(3)
plt.tight_layout()
plt.savefig('enthalpyofmixingAuPtfrominput.png', dpi=400, bboxinches='tight')
plt.show() ```
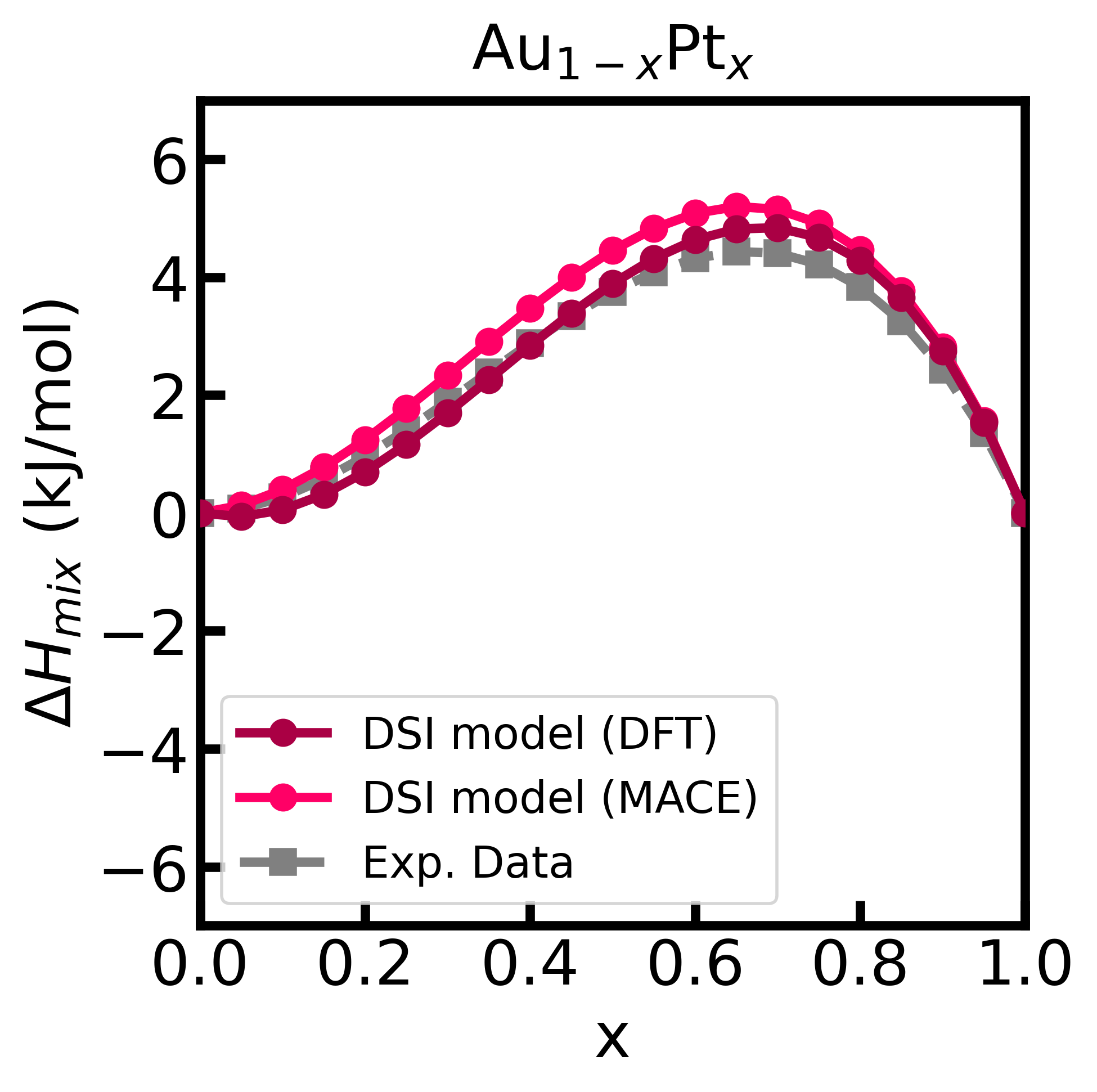
Figure 2 - Enthalpy of mixing of the Au-Pt alloy computed using the DSI model and energy matrix given from input (calculated with GPAW).
Polymorphism
Ternary alloy
Phase diagram (NOT WORKING)
By analyzing the mixing enthalpies and entropies, we can calculate the Gibbs free energy of the Au–Pt alloy mixture and determine both the spinodal and binodal (solvus) decomposition curves. These curves, which form key features of the alloy's phase diagram, delineate regions of differing stability: below the binodal curve, the solid solution (Au, Pt) is metastable, whereas it becomes unstable beneath the spinodal curve.
We begin by defining a temperature range over which to calculate the spinodal and binodal curves. Optionally, the results can be saved in CSV files. ```python
temperatures = np.arange(300, 3001, 5)
spinodal curve
dfspinodal = blendpy.getspinodaldecomposition(temperatures = temperatures, npoints = 501) dfspinodal.tocsv("data/phasediagram/spinodal_AuPt.csv", index=False, header=True, sep=',')
binodal curve
dfbinodal = blendpy.getbinodalcurve(temperatures = temperatures, npoints=501) dfbinodal.tocsv("data/phasediagram/binodal_AuPt.csv", index=False, header=True, sep=',') ```
To plot the phase diagram featuring the spinodal and binodal decomposition curves, we proceed as follows: ```python import pandas as pd
Create figure and axis
fig, ax = plt.subplots(1,1, figsize=(8,8))
x = np.linspace(0, 1, 101)
Configure axis labels and limits
ax.setxlabel("$x$", fontsize=20) ax.setylabel("$T$ (K)", fontsize=20) ax.setxlim(0,1) ax.setylim(300, 2500) ax.set_xticks(np.linspace(0,1,6))
Plot the data
ax.plot(dfspinodal['x'], dfspinodal['t'], color='#d53e4f', linestyle='--', linewidth=3, label="Spinodal curve") ax.plot(dfbinodal['x'], dfbinodal['t'], color='#d53e4f', linewidth=3, label="Binodal curve")
Fill below the curves with transparency (alpha=0.3 means 30% opacity)
ax.fillbetween(dfspinodal['x'], dfspinodal['t'], 300, color='#d53e4f', alpha=0.3) ax.fillbetween(dfbinodal['x'], dfbinodal['t'], 300, color='#d53e4f', alpha=0.3) ax.legend(loc="best", fontsize=20)
Add text annotations
ax.text(0.2, 1500, "Stable", fontsize=20, ha='center', va='center') ax.text(0.4, 950, "Metastable", fontsize=20, ha='center', va='center', rotation=60) ax.text(0.7, 700, "Unstable", fontsize=20, ha='center', va='center')
Customize tick parameters
ax.tickparams(direction='in', axis='both', which='major', labelsize=20, width=3, length=8) ax.setboxaspect(1) for spine in ax.spines.values(): spine.setlinewidth(3)
plt.tight_layout()
plt.savefig("phasediagram.png", dpi=600, format='png', bboxinches='tight')
plt.show() ```

Figure 2 - Phase diagram of the Au–Pt alloy computed using the DSI model and MACE interatomic potentials.
License
This is an open source code under MIT License.
Acknowledgements
We thank financial support from FAPESP (Grant No. 2022/14549-3), INCT Materials Informatics (Grant No. 406447/2022-5), and CNPq (Grant No. 311324/2020-7).
References
[^fn1]: Batatia, I., et al., MACE: Higher Order Equivariant Message Passing Neural Networks for Fast and Accurate Force Fields Adv. Neural Inf. Process Syst. 35, 11423 (2022).
[^fn2]: Mortensen, J. J., et al., GPAW: An open Python package for electronic structure calculations J. Chem. Phys. 160, 092503 (2024).
[^fn3]: Okamoto, H. and Massalski, T., The Au−Pt (Gold-Platinum) system Bull. Alloy Phase Diagr. 6, 46-56 (1985).
Owner
- Name: Leandro Seixas
- Login: leseixas
- Kind: user
- Location: São Paulo, Brazil
- Company: Mackenzie Presbyterian University
- Website: https://macksim.org
- Repositories: 15
- Profile: https://github.com/leseixas
Assistant Professor Doctor at Mackenzie Presbyterian University.
GitHub Events
Total
- Release event: 52
- Watch event: 2
- Push event: 147
- Create event: 52
Last Year
- Release event: 52
- Watch event: 2
- Push event: 147
- Create event: 52
Packages
- Total packages: 1
-
Total downloads:
- pypi 257 last-month
- Total dependent packages: 0
- Total dependent repositories: 0
- Total versions: 56
- Total maintainers: 1
pypi.org: blendpy
Blendpy is a computational toolkit for investigating thermodynamic models of alloys using first-principles calculations.
- Homepage: https://github.com/leseixas/blendpy
- Documentation: https://blendpy.readthedocs.io/
- License: MIT License Copyright (c) 2025 Leandro Seixas Rocha <leandro.fisica@gmail.com> Permission is hereby granted, free of charge, to any person obtaining a copy of this software and associated documentation files (the "Software"), to deal in the Software without restriction, including without limitation the rights to use, copy, modify, merge, publish, distribute, sublicense, and/or sell copies of the Software, and to permit persons to whom the Software is furnished to do so, subject to the following conditions: The above copyright notice and this permission notice shall be included in all copies or substantial portions of the Software. THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.
-
Latest release: 25.3.46
published 11 months ago



 -->
-->