insect-detect-ml
Notebooks for detection and classification model training. Insect classification model. Python scripts for processing of data, collected with the Insect Detect DIY camera trap.
Science Score: 67.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
✓CITATION.cff file
Found CITATION.cff file -
✓codemeta.json file
Found codemeta.json file -
✓.zenodo.json file
Found .zenodo.json file -
✓DOI references
Found 8 DOI reference(s) in README -
✓Academic publication links
Links to: zenodo.org -
○Academic email domains
-
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (11.2%) to scientific vocabulary
Keywords
Repository
Notebooks for detection and classification model training. Insect classification model. Python scripts for processing of data, collected with the Insect Detect DIY camera trap.
Basic Info
- Host: GitHub
- Owner: maxsitt
- License: agpl-3.0
- Language: Jupyter Notebook
- Default Branch: main
- Homepage: https://maxsitt.github.io/insect-detect-docs/
- Size: 52.4 MB
Statistics
- Stars: 18
- Watchers: 1
- Forks: 3
- Open Issues: 0
- Releases: 3
Topics
Metadata Files
README.md
Insect Detect ML - Model training and data processing
This repository contains Jupyter notebooks that can be used to train custom YOLOv5, YOLOv6, YOLOv7 and YOLOv8 object detection models or a custom YOLOv5 image classification model. All notebooks can be run in Google Colab, where you will have access to a free cloud GPU for fast training without special hardware requirements.
The Python script for classification
of the captured insect images is available in the custom yolov5
fork and can be used together with the provided
insect classification model.
Use the process_metadata.py
script for post-processing of metadata .csv files with classification results.
Model training
You can find more information about detection model training at the Insect Detect Docs 📑.
- YOLOv5 detection model training
- YOLOv6 detection model training
- YOLOv7 detection model training
- YOLOv8 detection model training
The PyTorch model weights can be converted to .blob format at tools.luxonis.com for on-device inference with the Luxonis OAK devices.
You can find more information about classification model training at the Insect Detect Docs 📑.
The notebook for classification model training includes export to ONNX format for faster CPU inference.
Classification
The modified classification script
in the custom yolov5 fork includes the following added options:
--sort-top1sort the classified images to folders with the predicted top1 class as folder name--sort-probsort images first by probability and then by top1 class (requires --sort-top1)--concat-csvconcatenate all metadata .csv files and append classification results to new columns
More information about deployment of the classification script can be found at the Insect Detect Docs 📑.
Classification model
| Model
(.onnx) | size
(pixels) | Top1 Accuracytest
| Precisiontest
| Recalltest
| F1 scoretest
|
| --------------------- | --------------------- | -------------------------- | ---------------------- | ------------------- | --------------------- |
| EfficientNet-B0 | 128 | 0.972 | 0.971 | 0.967 | 0.969 |
Table Notes
- The model was trained to 20 epochs with image size 128, batch size 64 and default settings and hyperparameters. Reproduce the model training with the provided Google Colab notebook.
- Trained on Insect Detect - insect classification dataset v2 with 27 classes. To reproduce the dataset split, keep the default settings in the Colab notebook (train/val/test ratio = 0.7/0.2/0.1, random seed = 1).
- Dataset can be explored at Roboflow Universe. Export from Roboflow compresses the images and can lead to a decreased model accuracy. It is recommended to use the uncompressed dataset from Zenodo.
Full model metrics on dataset test split (click to expand)
| Class | Images | Top1 Accuracytest| Precisiontest
| Recalltest
| F1 scoretest
| | ------------ | ------ | -------------------------- | ---------------------- | ------------------- | --------------------- | | all | 2125 | 0.972 | 0.971 | 0.967 | 0.969 | | ant | 111 | 1.0 | 0.991 | 1.0 | 0.996 | | bee | 107 | 0.963 | 0.972 | 0.963 | 0.967 | | bee_apis | 31 | 1.0 | 0.969 | 1.0 | 0.984 | | bee_bombus | 127 | 1.0 | 0.992 | 1.0 | 0.996 | | beetle | 52 | 0.885 | 0.92 | 0.885 | 0.902 | | beetle_cocci | 78 | 0.987 | 1.0 | 0.987 | 0.994 | | beetle_oedem | 21 | 0.905 | 0.905 | 0.905 | 0.905 | | bug | 39 | 0.846 | 1.0 | 0.846 | 0.917 | | bug_grapho | 19 | 1.0 | 1.0 | 1.0 | 1.0 | | fly | 173 | 0.971 | 0.944 | 0.971 | 0.957 | | fly_empi | 19 | 1.0 | 1.0 | 1.0 | 1.0 | | fly_sarco | 33 | 0.909 | 0.938 | 0.909 | 0.923 | | fly_small | 167 | 0.958 | 0.952 | 0.958 | 0.955 | | hfly_episyr | 253 | 0.996 | 0.996 | 0.996 | 0.996 | | hfly_eristal | 197 | 0.99 | 0.995 | 0.99 | 0.992 | | hfly_eupeo | 137 | 0.985 | 0.993 | 0.985 | 0.989 | | hfly_myathr | 60 | 1.0 | 1.0 | 1.0 | 1.0 | | hfly_sphaero | 39 | 0.974 | 1.0 | 0.974 | 0.987 | | hfly_syrphus | 50 | 0.98 | 1.0 | 0.98 | 0.99 | | lepi | 24 | 1.0 | 0.96 | 1.0 | 0.98 | | none_bg | 86 | 0.988 | 0.966 | 0.988 | 0.977 | | none_bird | 8 | 1.0 | 1.0 | 1.0 | 1.0 | | none_dirt | 85 | 0.976 | 0.902 | 0.976 | 0.938 | | none_shadow | 66 | 0.924 | 0.953 | 0.924 | 0.938 | | other | 79 | 0.861 | 0.883 | 0.861 | 0.872 | | scorpionfly | 12 | 1.0 | 1.0 | 1.0 | 1.0 | | wasp | 52 | 1.0 | 1.0 | 1.0 | 1.0 |
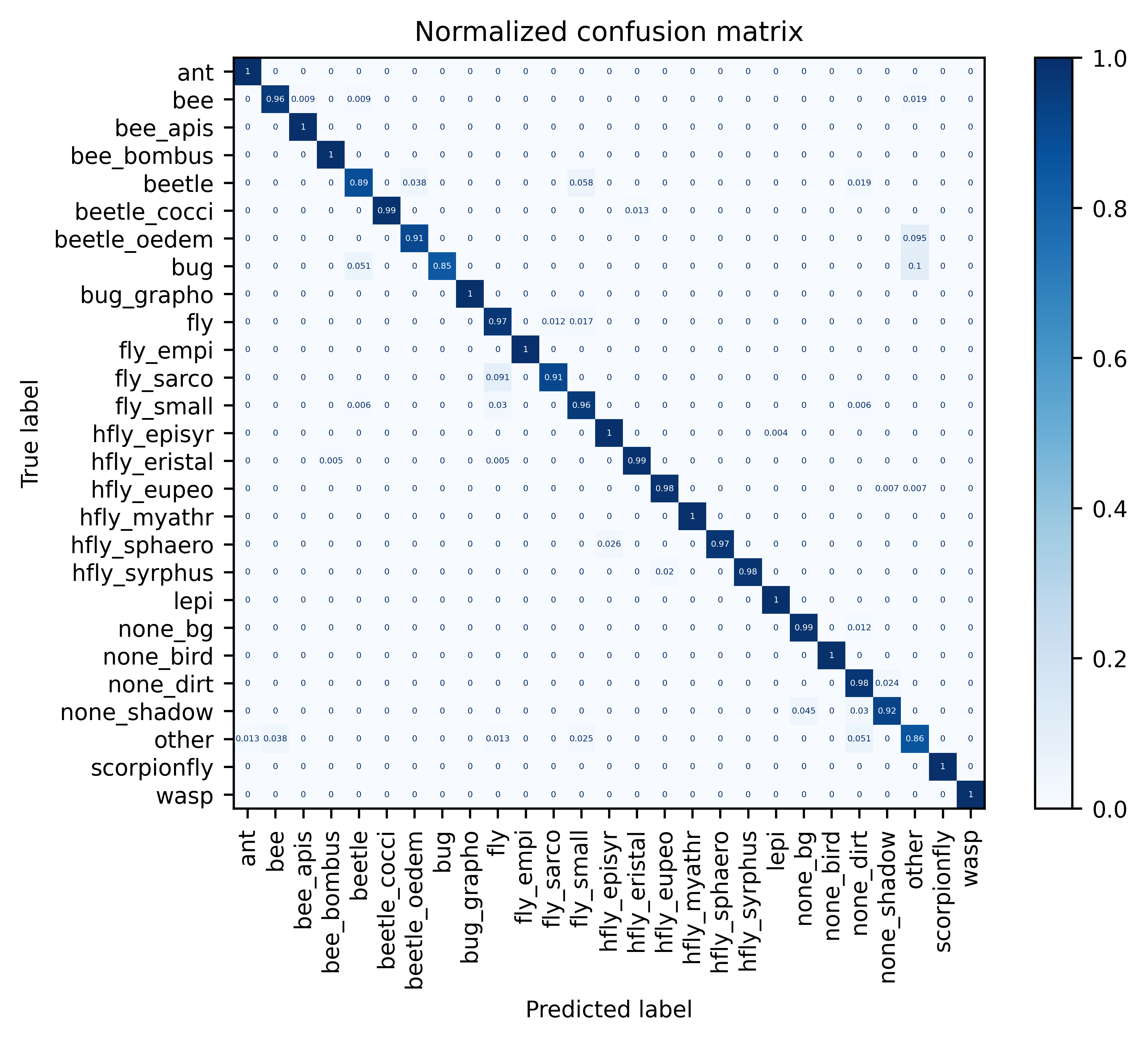
Full model metrics on dataset validation split (click to expand)
| Class | Images | Top1 Accuracyval| Precisionval
| Recallval
| F1 scoreval
| | ------------ | ------ | ------------------------- | --------------------- | ------------------ | -------------------- | | all | 4189 | 0.98 | 0.979 | 0.974 | 0.976 | | ant | 219 | 0.995 | 0.995 | 0.995 | 0.995 | | bee | 212 | 0.967 | 0.958 | 0.967 | 0.962 | | bee_apis | 58 | 1.0 | 0.967 | 1.0 | 0.983 | | bee_bombus | 252 | 1.0 | 0.996 | 1.0 | 0.998 | | beetle | 104 | 0.933 | 0.942 | 0.933 | 0.937 | | beetle_cocci | 155 | 1.0 | 1.0 | 1.0 | 1.0 | | beetle_oedem | 39 | 0.897 | 0.972 | 0.897 | 0.933 | | bug | 78 | 0.949 | 0.961 | 0.949 | 0.955 | | bug_grapho | 37 | 1.0 | 1.0 | 1.0 | 1.0 | | fly | 343 | 0.983 | 0.939 | 0.983 | 0.96 | | fly_empi | 35 | 1.0 | 0.972 | 1.0 | 0.986 | | fly_sarco | 63 | 0.841 | 0.964 | 0.841 | 0.898 | | fly_small | 332 | 0.97 | 0.982 | 0.97 | 0.976 | | hfly_episyr | 503 | 0.996 | 0.996 | 0.996 | 0.996 | | hfly_eristal | 390 | 1.0 | 1.0 | 1.0 | 1.0 | | hfly_eupeo | 271 | 0.989 | 0.993 | 0.989 | 0.991 | | hfly_myathr | 118 | 0.992 | 1.0 | 0.992 | 0.996 | | hfly_sphaero | 74 | 1.0 | 0.987 | 1.0 | 0.993 | | hfly_syrphus | 97 | 1.0 | 0.99 | 1.0 | 0.995 | | lepi | 45 | 0.978 | 0.978 | 0.978 | 0.978 | | none_bg | 170 | 0.988 | 0.982 | 0.988 | 0.985 | | none_bird | 13 | 1.0 | 1.0 | 1.0 | 1.0 | | none_dirt | 167 | 0.982 | 0.976 | 0.982 | 0.979 | | none_shadow | 129 | 0.969 | 0.984 | 0.969 | 0.977 | | other | 158 | 0.88 | 0.903 | 0.88 | 0.891 | | scorpionfly | 24 | 1.0 | 1.0 | 1.0 | 1.0 | | wasp | 103 | 0.99 | 1.0 | 0.99 | 0.995 |

Metadata post-processing
Install the required packages by running:
python.exe -m pip install -r requirements.txt
Or use the Python Launcher for Windows with:
py -m pip install -r requirements.txt
The process_metadata.py
script can be used to automatically post-process the concatenated metadata .csv file after the
classification
step, as it will still contain multiple rows for each tracked insect.
The output of the script includes a *top1_final.csv file in which each row
corresponds to an individual tracked insect and its classification result
with the highest weighted probability. Additionally, several
plots
are generated that can give a first overview of the processed metadata.
More information about deployment of the post-processing script can be found at the Insect Detect Docs 📑.
Image processing
The process_images.py
script can be used to calculate different metrics of the captured images
(e.g. mean/median/min/max width/height) and remove corrupted .jpg images
from the data folder (camera trap output). These can be rarely generated
by the automated monitoring script (e.g. in power outage situations) and
will cause an error while running the classification script.
License
This repository is licensed under the terms of the GNU Affero General Public License v3.0 (GNU AGPLv3).
Citation
If you use resources from this repository, please cite our paper:
Sittinger M, Uhler J, Pink M, Herz A (2024) Insect detect: An open-source DIY camera trap for automated insect monitoring. PLoS ONE 19(4): e0295474. https://doi.org/10.1371/journal.pone.0295474
Owner
- Name: Maximilian Sittinger
- Login: maxsitt
- Kind: user
- Location: Germany
- Company: JKI
- Twitter: M_Sittinger
- Repositories: 2
- Profile: https://github.com/maxsitt
Citation (CITATION.cff)
cff-version: 1.2.0
message: "If you use resources from this repository, please cite it using this metadata."
title: "Insect Detect ML - Software for classification of images and analysis of metadata from a DIY camera trap system"
authors:
- family-names: Sittinger
given-names: Maximilian
orcid: "https://orcid.org/0000-0002-4096-8556"
license: AGPL-3.0
doi: 10.5281/zenodo.7502195
repository-code: "https://github.com/maxsitt/insect-detect-ml"
url: "https://maxsitt.github.io/insect-detect-docs/"
version: 1.2
date-released: "2023-06-08"
preferred-citation:
type: article
authors:
- family-names: Sittinger
given-names: Maximilian
- family-names: Uhler
given-names: Johannes
- family-names: Pink
given-names: Maximilian
- family-names: Herz
given-names: Annette
title: "Insect detect: An open-source DIY camera trap for automated insect monitoring"
journal: "PLOS ONE"
year: 2024
month: 4
volume: 19
issue: 4
start: 1
end: 28
doi: 10.1371/journal.pone.0295474
url: "https://doi.org/10.1371/journal.pone.0295474"
GitHub Events
Total
- Watch event: 3
- Fork event: 1
Last Year
- Watch event: 3
- Fork event: 1
Dependencies
- Pillow *
- distinctipy *
- matplotlib *
- pandas *

