HNN-core
HNN-core: A Python software for cellular and circuit-level interpretation of human MEG/EEG - Published in JOSS (2023)
Science Score: 100.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
✓CITATION.cff file
Found CITATION.cff file -
✓codemeta.json file
Found codemeta.json file -
✓.zenodo.json file
Found .zenodo.json file -
✓DOI references
Found 8 DOI reference(s) in README and JOSS metadata -
✓Academic publication links
Links to: joss.theoj.org -
✓Committers with academic emails
5 of 31 committers (16.1%) from academic institutions -
○Institutional organization owner
-
✓JOSS paper metadata
Published in Journal of Open Source Software
Keywords
Keywords from Contributors
Scientific Fields
Repository
Simulation and optimization of neural circuits for MEG/EEG source estimates
Basic Info
- Host: GitHub
- Owner: jonescompneurolab
- License: bsd-3-clause
- Language: Python
- Default Branch: master
- Homepage: https://jonescompneurolab.github.io/hnn-core/
- Size: 172 MB
Statistics
- Stars: 62
- Watchers: 11
- Forks: 68
- Open Issues: 141
- Releases: 8
Topics
Metadata Files
README.md
hnn-core
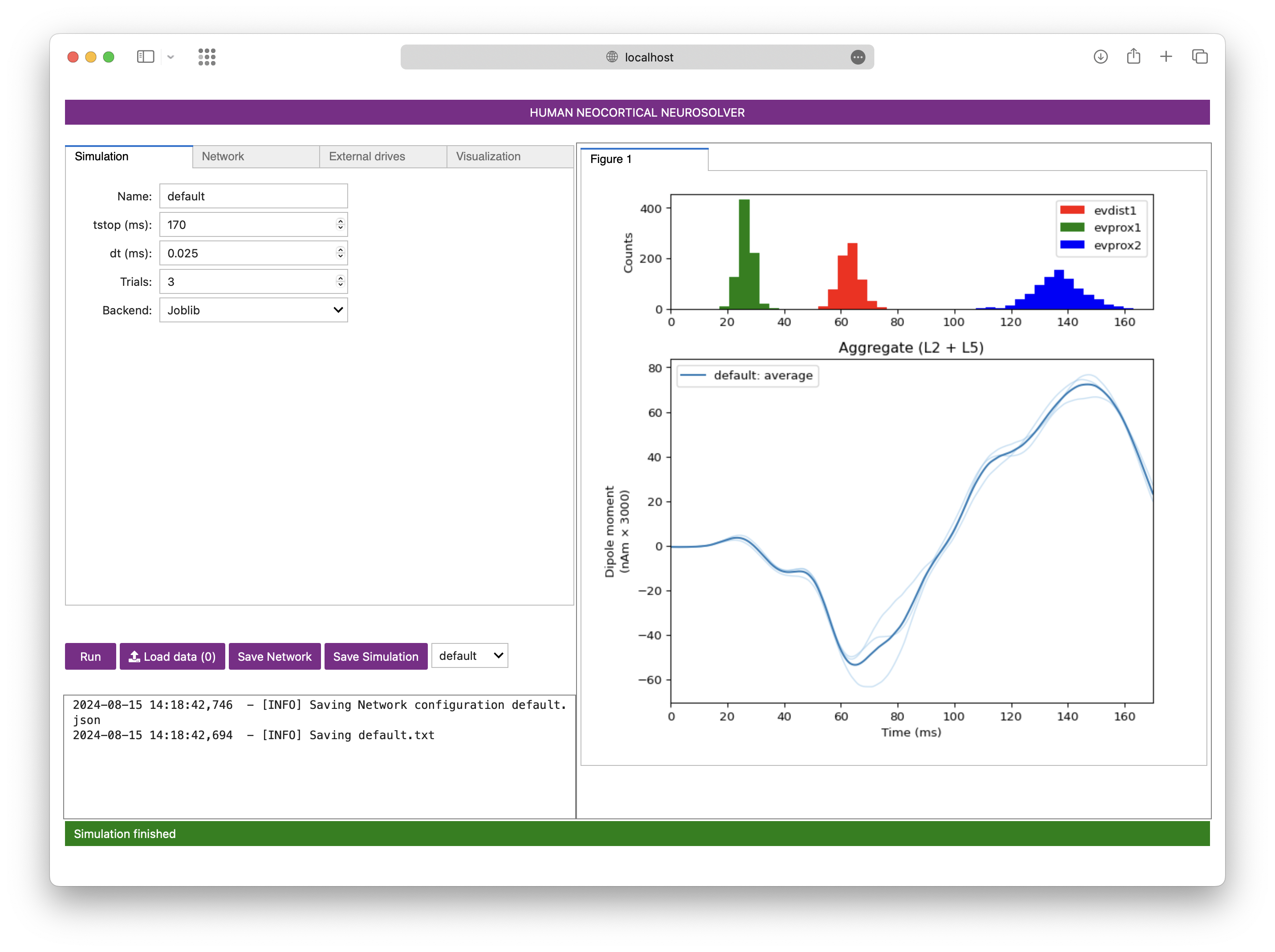
About
This is a leaner and cleaner version of the code based off the HNN repository.
The Human Neocortical Neurosolver (HNN) is an open-source neural modeling tool designed to help researchers/clinicians interpret human brain imaging data. Based off the original HNN repository, HNN-core provides a convenient way to run simulations of an anatomically and biophysically detailed dynamical system model of human thalamocortical brain circuits with only a few lines of code. Given its modular, object-oriented design, HNN-core makes it easy to generate and evaluate hypotheses on the mechanistic origin of signals measured with magnetoencephalography (MEG), electroencephalography (EEG), or intracranial electrocorticography (ECoG). A unique feature of the HNN model is that it accounts for the biophysics generating the primary electric currents underlying such data, so simulation results are directly comparable to source localized data (current dipoles in units of nano-Ampere-meters); this enables precise tuning of model parameters to match characteristics of recorded signals. Multimodal neurophysiology data such as local field potential (LFP), current-source density (CSD), and spiking dynamics can also be simulated simultaneously with current dipoles.
While the HNN-core API is designed to be flexible and serve users with varying levels of coding expertise, the HNN-core GUI is designed to be useful to researchers with no formal computational neural modeling or coding experience.
For more information visit https://hnn.brown.edu. There, we describe the use of HNN in studying the circuit-level origin of some of the most commonly measured MEG/EEG and ECoG signals: event related potentials (ERPs) and low frequency rhythms (alpha/beta/gamma).
Contributors are very welcome! Please read our Contributing Guide if you are interested.
Please consider supporting HNN development efforts by voluntarily providing your demographic information here! Note that any demographic information we collect is anonymized and aggregated for reporting on the grants that fund the continued development of HNN. All questions are voluntary.
Installation
See Installation Guide. To install hnn-core with the minimum dependencies
on Mac or Linux, simply do:
$ pip install hnn_core
hnn-core currently supports Python 3.9 through 3.13, inclusively.
If you want to track the latest developments of hnn-core, you can
install the current version of the code (nightly) with:
$ pip install --upgrade https://api.github.com/repos/jonescompneurolab/hnn-core/zipball/master
If you are interested in features like GUI, Optimization, or Parallel support, or are on Windows, then please see our Installation Guide.
Documentation and examples
Once you have installed hnn_core and the dependencies for the features you
want, we recommend downloading and executing the example
scripts
provided on the documentation
pages (as well as in the
GitHub repository).
Note that python plots are by default non-interactive (blocking): each
plot must thus be closed before the code execution continues. We
recommend using and 'interactive' python interpreter such as
ipython:
$ ipython --matplotlib
and executing the scripts using the %run-magic:
%run plot_simulate_evoked.py
When executed in this manner, the scripts will execute entirely, after which all plots will be shown. For an even more interactive experience, in which you execute code and interrogate plots in sequential blocks, we recommend editors such as VS Code and Spyder.
Bug reports
Use the GitHub Issues tracker to report bugs. For user questions and scientific discussions, please see our GitHub Discussions page.
Interested in Contributing?
Please read our Contributing Guide and make sure to abide by our Code of Conduct.
Governance Structure
Our governance structure can be found here.
Citing
If you use HNN-core in your work, please cite our publication in JOSS:
Jas et al., (2023). HNN-core: A Python software for cellular and circuit-level interpretation of human MEG/EEG. Journal of Open Source Software, 8(92), 5848, https://doi.org/10.21105/joss.05848
Owner
- Name: jonescompneurolab
- Login: jonescompneurolab
- Kind: organization
- Repositories: 6
- Profile: https://github.com/jonescompneurolab
JOSS Publication
HNN-core: A Python software for cellular and circuit-level interpretation of human MEG/EEG
Authors

Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Boston, MA, USA

Department of Neuroscience, Brown University, Providence, RI, USA, Robert J. and Nancy D. Carney Institute for Brain Science, Brown University, Providence, RI, USA

Department of Neuroscience, Brown University, Providence, RI, USA, Robert J. and Nancy D. Carney Institute for Brain Science, Brown University, Providence, RI, USA
Department of Neuroscience, Brown University, Providence, RI, USA, Robert J. and Nancy D. Carney Institute for Brain Science, Brown University, Providence, RI, USA
Department of Psychological and Brain Sciences, Indiana University Bloomington, Bloomington, IN, USA

Robert J. and Nancy D. Carney Institute for Brain Science, Brown University, Providence, RI, USA

Department of Biomedical Engineering, University of Miami, Coral Gables, FL, USA
Department of Psychiatry and Behavioral Health, Penn State Milton S. Hershey Medical Center, Penn State College of Medicine, Hershey, PA, USA

Robert J. and Nancy D. Carney Institute for Brain Science, Brown University, Providence, RI, USA

Department of Psychology, University of Jyväskylä, Jyväskylä, Finland, Jyväskylä Centre for Interdisciplinary Brain Research, University of Jyväskylä, Jyväskylä, Finland

Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Boston, MA, USA, Department of Radiology, Harvard Medical School, Boston, MA, USA
Robert J. and Nancy D. Carney Institute for Brain Science, Brown University, Providence, RI, USA, Department of Molecular and Cell Biology; Innovative Genomics Institute, University of California, Berkeley, Berkeley, CA, USA

Center for Biomedical Imaging and Neuromodulation, Nathan S. Kline Institute for Psychiatric Research, Orangeburg, NY, USA, Department of Psychiatry, New York University Grossman School of Medicine, New York, NY, USA

Department of Computer Science and Engineering, National Institute of Technology Karnataka, Karnataka, India
Robert J. and Nancy D. Carney Institute for Brain Science, Brown University, Providence, RI, USA, Florida State University, Tallahassee, FL, USA

Department of Psychiatry and Human Behavior, Brown University, Providence, RI, USA, Rhode Island Hospital, Providence, RI, USA
Tags
neuroscience EEG MEG modeling neocortexCitation (CITATION.cff)
cff-version: "1.2.0"
authors:
- family-names: Jas
given-names: Mainak
orcid: "https://orcid.org/0000-0002-3199-9027"
- family-names: Thorpe
given-names: Ryan
orcid: "https://orcid.org/0000-0003-2491-8599"
- family-names: Tolley
given-names: Nicholas
orcid: "https://orcid.org/0000-0003-0358-0074"
- family-names: Bailey
given-names: Christopher
orcid: "https://orcid.org/0000-0003-3318-3344"
- family-names: Brandt
given-names: Steven
- family-names: Caldwell
given-names: Blake
orcid: "https://orcid.org/0000-0002-6882-6998"
- family-names: Cheng
given-names: Huzi
- family-names: Daniels
given-names: Dylan
orcid: "https://orcid.org/0009-0008-1958-353X"
- family-names: Pujol
given-names: Carolina Fernandez
orcid: "https://orcid.org/0009-0003-0611-1270"
- family-names: Khalil
given-names: Mostafa
- family-names: Kanekar
given-names: Samika
orcid: "https://orcid.org/0000-0002-6111-4461"
- family-names: Kohl
given-names: Carmen
orcid: "https://orcid.org/0000-0001-7585-595X"
- family-names: Kolozsvári
given-names: Orsolya
orcid: "https://orcid.org/0000-0002-1619-6314"
- family-names: Lankinen
given-names: Kaisu
orcid: "https://orcid.org/0000-0003-2210-2385"
- family-names: Loi
given-names: Kenneth
- family-names: Neymotin
given-names: Sam
orcid: "https://orcid.org/0000-0003-3646-5195"
- family-names: Partani
given-names: Rajat
orcid: "https://orcid.org/0000-0002-6863-7046"
- family-names: Pelah
given-names: Mattan
- family-names: Rockhill
given-names: Alex
orcid: "https://orcid.org/0000-0003-3868-7453"
- family-names: Sherif
given-names: Mohamed
orcid: "https://orcid.org/0000-0002-8951-1645"
- family-names: Hamalainen
given-names: Matti
orcid: "https://orcid.org/0000-0001-6841-112X"
- family-names: Jones
given-names: Stephanie
orcid: "https://orcid.org/0000-0001-6760-5301"
contact:
- family-names: Jones
given-names: Stephanie
orcid: "https://orcid.org/0000-0001-6760-5301"
doi: 10.5281/zenodo.10289164
message: If you use this software, please cite our article in the
Journal of Open Source Software.
preferred-citation:
authors:
- family-names: Jas
given-names: Mainak
orcid: "https://orcid.org/0000-0002-3199-9027"
- family-names: Thorpe
given-names: Ryan
orcid: "https://orcid.org/0000-0003-2491-8599"
- family-names: Tolley
given-names: Nicholas
orcid: "https://orcid.org/0000-0003-0358-0074"
- family-names: Bailey
given-names: Christopher
orcid: "https://orcid.org/0000-0003-3318-3344"
- family-names: Brandt
given-names: Steven
- family-names: Caldwell
given-names: Blake
orcid: "https://orcid.org/0000-0002-6882-6998"
- family-names: Cheng
given-names: Huzi
- family-names: Daniels
given-names: Dylan
orcid: "https://orcid.org/0009-0008-1958-353X"
- family-names: Pujol
given-names: Carolina Fernandez
orcid: "https://orcid.org/0009-0003-0611-1270"
- family-names: Khalil
given-names: Mostafa
- family-names: Kanekar
given-names: Samika
orcid: "https://orcid.org/0000-0002-6111-4461"
- family-names: Kohl
given-names: Carmen
orcid: "https://orcid.org/0000-0001-7585-595X"
- family-names: Kolozsvári
given-names: Orsolya
orcid: "https://orcid.org/0000-0002-1619-6314"
- family-names: Lankinen
given-names: Kaisu
orcid: "https://orcid.org/0000-0003-2210-2385"
- family-names: Loi
given-names: Kenneth
- family-names: Neymotin
given-names: Sam
orcid: "https://orcid.org/0000-0003-3646-5195"
- family-names: Partani
given-names: Rajat
orcid: "https://orcid.org/0000-0002-6863-7046"
- family-names: Pelah
given-names: Mattan
- family-names: Rockhill
given-names: Alex
orcid: "https://orcid.org/0000-0003-3868-7453"
- family-names: Sherif
given-names: Mohamed
orcid: "https://orcid.org/0000-0002-8951-1645"
- family-names: Hamalainen
given-names: Matti
orcid: "https://orcid.org/0000-0001-6841-112X"
- family-names: Jones
given-names: Stephanie
orcid: "https://orcid.org/0000-0001-6760-5301"
date-published: 2023-12-15
doi: 10.21105/joss.05848
issn: 2475-9066
issue: 92
journal: Journal of Open Source Software
publisher:
name: Open Journals
start: 5848
title: "HNN-core: A Python software for cellular and circuit-level
interpretation of human MEG/EEG"
type: article
url: "https://joss.theoj.org/papers/10.21105/joss.05848"
volume: 8
title: "HNN-core: A Python software for cellular and circuit-level
interpretation of human MEG/EEG"
GitHub Events
Total
- Fork event: 15
- Create event: 16
- Release event: 1
- Issues event: 95
- Watch event: 6
- Delete event: 8
- Member event: 1
- Issue comment event: 388
- Push event: 166
- Gollum event: 25
- Pull request review comment event: 255
- Pull request review event: 264
- Pull request event: 176
Last Year
- Fork event: 15
- Create event: 16
- Release event: 1
- Issues event: 95
- Watch event: 6
- Delete event: 8
- Member event: 1
- Issue comment event: 388
- Push event: 166
- Gollum event: 25
- Pull request review comment event: 255
- Pull request review event: 264
- Pull request event: 176
Committers
Last synced: 7 months ago
Top Committers
| Name | Commits | |
|---|---|---|
| Mainak Jas | m****s@g****m | 656 |
| Nick Tolley | n****y@g****m | 565 |
| George Dang | 5****g | 350 |
| Ryan Thorpe | r****e@b****u | 343 |
| Huzi Cheng | t****s@i****m | 212 |
| Christopher J. Bailey | b****j@g****m | 202 |
| Blake Caldwell | 2****l@g****m | 139 |
| Camilo Diaz | k****9@g****m | 93 |
| Carolina Fernandez | c****8@m****u | 80 |
| Austin E. Soplata | a****a | 63 |
| katduecker | k****r@g****m | 32 |
| samadpls | a****1@g****m | 32 |
| raj1701 | r****i@g****m | 32 |
| dylansdaniels | d****s@g****m | 21 |
| kenloi | k****i@b****u | 19 |
| Carmen | k****1@g****m | 15 |
| mohdsherif | m****b@g****m | 14 |
| spbrandt | 1****t | 10 |
| Orsolya Beatrix Kolozsvari | o****i@g****m | 7 |
| Kaisu Lankinen | k****n@m****u | 6 |
| Samika Kanekar | s****n@g****m | 6 |
| Stephanie R. Jones | S****s@B****u | 5 |
| Alex | a****l@m****g | 5 |
| Yaroslav Halchenko | d****n@o****m | 5 |
| Maira Usman | m****3@g****m | 3 |
| Tianqi Cheng | t****g@T****l | 3 |
| Dikshant jha | 1****4 | 1 |
| Pynmash | 1****h | 1 |
| Shehroz Kashif | 1****f | 1 |
| mjpelah | 9****h | 1 |
| and 1 more... | ||
Committer Domains (Top 20 + Academic)
Issues and Pull Requests
Last synced: 6 months ago
All Time
- Total issues: 154
- Total pull requests: 336
- Average time to close issues: 9 months
- Average time to close pull requests: 25 days
- Total issue authors: 15
- Total pull request authors: 24
- Average comments per issue: 1.61
- Average comments per pull request: 2.34
- Merged pull requests: 218
- Bot issues: 9
- Bot pull requests: 2
Past Year
- Issues: 83
- Pull requests: 208
- Average time to close issues: 25 days
- Average time to close pull requests: 13 days
- Issue authors: 8
- Pull request authors: 16
- Average comments per issue: 0.7
- Average comments per pull request: 1.86
- Merged pull requests: 128
- Bot issues: 9
- Bot pull requests: 0
Top Authors
Issue Authors
- asoplata (51)
- gtdang (34)
- jasmainak (17)
- github-actions[bot] (9)
- ntolley (9)
- kmilo9999 (7)
- dylansdaniels (6)
- rythorpe (5)
- darcywaller (4)
- blakecaldwell (4)
- katduecker (3)
- samadpls (2)
- chenghuzi (1)
- kenneth59715 (1)
- hanbuck30 (1)
Pull Request Authors
- asoplata (120)
- gtdang (67)
- dylansdaniels (26)
- kmilo9999 (21)
- ntolley (18)
- samadpls (14)
- dikshant182004 (10)
- katduecker (9)
- jasmainak (9)
- Myrausman (6)
- Chetank99 (5)
- carolinafernandezp (5)
- Freedisch (4)
- pynmash (4)
- Shehrozkashif (4)
Top Labels
Issue Labels
Pull Request Labels
Packages
- Total packages: 1
-
Total downloads:
- pypi 87 last-month
- Total dependent packages: 0
- Total dependent repositories: 3
- Total versions: 11
- Total maintainers: 3
pypi.org: hnn-core
Code for biophysical simulation of a cortical column using Neuron
- Documentation: https://hnn-core.readthedocs.io/
- License: BSD (3-clause)
-
Latest release: 0.4.2
published 8 months ago
Rankings
Dependencies
- actions/checkout v2 composite
- conda-incubator/setup-miniconda v2 composite
- actions/checkout v2 composite
- conda-incubator/setup-miniconda v2 composite
- actions/checkout v2 composite
- conda-incubator/setup-miniconda v2 composite
- jupyter/minimal-notebook 65761486d5d3 build
- NEURON *
- ipywidgets *
- matplotlib *
- scipy *
- voila *
- NEURON *
- h5io *
- matplotlib >=3.5.3
- numpy *
- scipy *

