https://github.com/brainglobe/brainreg-napari
Automated 3D brain registration in napari with support for multiple species and atlases.
Science Score: 51.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
✓CITATION.cff file
Found CITATION.cff file -
○codemeta.json file
-
○.zenodo.json file
-
✓DOI references
Found 8 DOI reference(s) in README -
✓Academic publication links
Links to: nature.com -
✓Committers with academic emails
1 of 6 committers (16.7%) from academic institutions -
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (15.3%) to scientific vocabulary
Keywords
Keywords from Contributors
Repository
Automated 3D brain registration in napari with support for multiple species and atlases.
Basic Info
- Host: GitHub
- Owner: brainglobe
- License: mit
- Language: Python
- Default Branch: main
- Homepage: https://brainglobe.info/documentation/brainreg/index.html
- Size: 116 KB
Statistics
- Stars: 16
- Watchers: 4
- Forks: 6
- Open Issues: 0
- Releases: 4
Topics
Metadata Files
README.md
THIS PACKAGE HAS MOVED
As of the release of brainreg version 1.0.0, brainreg-napari is now a part of brainreg.
If you are looking to install the brainglobe-napari plugin, please install brainreg with it's optional napari dependency as detailed in the installation instructions on the website or repository.
Before you update, you should also remove the old brainreg-napari package from your environment using either
bash
python -m pip uninstall brainreg-napari # If you installed via pip
conda remove brainreg-napari # If you installed via conda
You can find the old documentation and installation instructions below, but please note this version of the package should be considered unmaintained.
brainreg-napari
Napari plugin to run brainreg, developed by Stephen Lenzi.
Installation
bash
pip install brainreg-napari
Usage
Documentation and tutorials for the plugin can be found here.
For segmentation of bulk structures in 3D space (e.g. injection sites, Neuropixels probes), please see brainreg-segment.
This software is at a very early stage, and was written with our data in mind. Over time we hope to support other data types/formats. If you have any issues, please get in touch on the forum or by raising an issue.
Details
brainreg is an update to amap (itself a port of the original Java software) to include multiple registration backends, and to support the many atlases provided by bg-atlasapi.
The aim of brainreg is to register the template brain (e.g. from the Allen Reference Atlas) to the sample image. Once this is complete, any other image in the template space can be aligned with the sample (such as region annotations, for segmentation of the sample image). The template to sample transformation can also be inverted, allowing sample images to be aligned in a common coordinate space.
To do this, the template and sample images are filtered, and then registered in a three step process (reorientation, affine registration, and freeform registration.) The resulting transform from template to standard space is then applied to the atlas.
Full details of the process are in the
original aMAP paper.
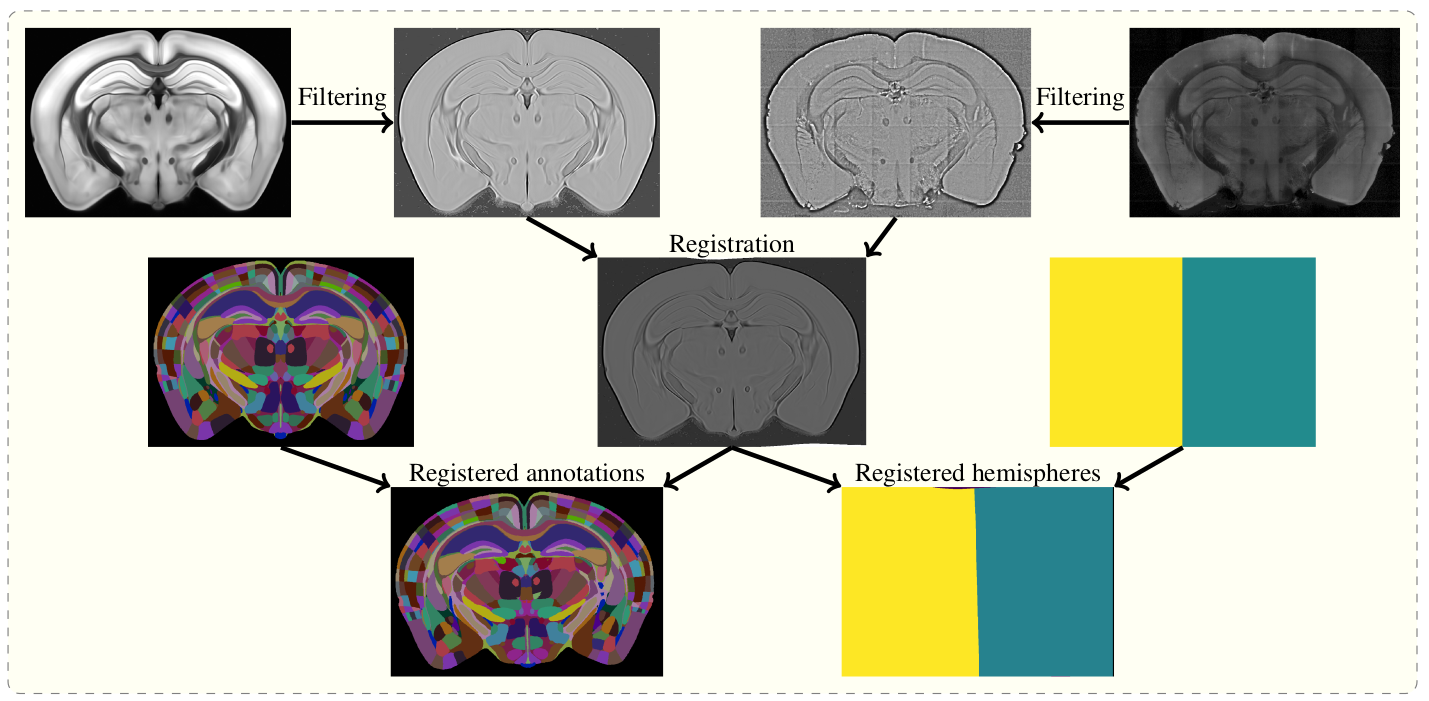 Overview of the registration process
Overview of the registration process
Contributing
Contributions to brainreg-napari are more than welcome. Please see the developers guide.
Citing brainreg
If you find brainreg useful, and use it in your research, please let us know and also cite the paper:
Tyson, A. L., Vélez-Fort, M., Rousseau, C. V., Cossell, L., Tsitoura, C., Lenzi, S. C., Obenhaus, H. A., Claudi, F., Branco, T., Margrie, T. W. (2022). Accurate determination of marker location within whole-brain microscopy images. Scientific Reports, 12, 867 doi.org/10.1038/s41598-021-04676-9
Please also cite aMAP (the original pipeline from which this software is based):
Niedworok, C.J., Brown, A.P.Y., Jorge Cardoso, M., Osten, P., Ourselin, S., Modat, M. and Margrie, T.W., (2016). AMAP is a validated pipeline for registration and segmentation of high-resolution mouse brain data. Nature Communications. 7, 1–9. https://doi.org/10.1038/ncomms11879
Lastly, if you can, please cite the BrainGlobe Atlas API that provided the atlas:
Claudi, F., Petrucco, L., Tyson, A. L., Branco, T., Margrie, T. W. and Portugues, R. (2020). BrainGlobe Atlas API: a common interface for neuroanatomical atlases. Journal of Open Source Software, 5(54), 2668, https://doi.org/10.21105/joss.02668
Don't forget to cite the developers of the atlas that you used (e.g. the Allen Brain Atlas)!
Owner
- Name: BrainGlobe
- Login: brainglobe
- Kind: organization
- Location: London/Munich
- Website: https://brainglobe.info/
- Twitter: brain_globe
- Repositories: 28
- Profile: https://github.com/brainglobe
Open python tools for morphological analyses in systems neuroscience
Citation (CITATION.CFF)
cff-version: 1.2.0
message: If you use this software, please cite it using these metadata.
title: brainreg-napari
authors:
- family-names: "Tyson"
given-names: "A."
- family-names: "Vélez-Fort"
given-names: "M."
- family-names: "Rousseau"
given-names: "C."
- family-names: "Cossell"
given-names: "L."
- family-names: "Tsitoura"
given-names: " C."
- family-names: "Lenzi"
given-names: " S."
- family-names: "Obenhaus"
given-names: "H."
- family-names: "Claudi"
given-names: "F."
- family-names: "Branco"
given-names: "T."
- family-names: "Margrie"
given-names: "T."
url: https://github.com/brainglobe/brainreg-napari
preferred-citation:
type: article
title: Accurate determination of marker location within whole-brain microscopy images
journal: Scientific Reports
volume: 12
pages: 867
doi: 10.1038/s41598-021-04676-9
year: 2022
date-published: 2022-01-18
authors:
- family-names: "Tyson"
given-names: "A."
- family-names: "Vélez-Fort"
given-names: "M."
- family-names: "Rousseau"
given-names: "C."
- family-names: "Cossell"
given-names: "L."
- family-names: "Tsitoura"
given-names: " C."
- family-names: "Lenzi"
given-names: "S."
- family-names: "Obenhaus"
given-names: "H."
- family-names: "Claudi"
given-names: "F."
- family-names: "Branco"
given-names: "T."
- family-names: "Margrie"
given-names: "T."
references:
- type: article
title: AMAP is a validated pipeline for registration and segmentation of high-resolution mouse brain data
journal: Nature Communications
volume: 7
pages: 1-9
doi: 10.1038/ncomms11879
year: 2016
date-published: 2016-07-07
authors:
- family-names: "Niedworok"
given-names: "C."
- family-names: "Brown"
given-names: "A."
- family-names: "Jorge"
given-names: "M."
- family-names: "Osten"
given-names: "P."
- family-names: "Ourselin"
given-names: "S."
- family-names: "Modat"
given-names: "M."
- family-names: "Margrie"
given-names: "T."
references:
- type: article
title: BrainGlobe Atlas API a common interface for neuroanatomical atlases
journal: Journal of Open Source Software
volume: 5
pages: 2668
doi: 10.21105/joss.02668
year: 2020
date-published: 2020-09-04
authors:
- family-names: "Claudi"
given-names: "F."
- family-names: "Petrucco"
given-names: "L."
- family-names: "Tyson"
given-names: "A."
- family-names: "Branco"
given-names: "T."
- family-names: "Margrie"
given-names: "T."
- family-names: "Portugues"
given-names: "R."
GitHub Events
Total
- Watch event: 1
Last Year
- Watch event: 1
Committers
Last synced: almost 3 years ago
All Time
- Total Commits: 82
- Total Committers: 6
- Avg Commits per committer: 13.667
- Development Distribution Score (DDS): 0.634
Top Committers
| Name | Commits | |
|---|---|---|
| David Stansby | d****y@g****m | 30 |
| stephen | s****i@g****m | 29 |
| Adam Tyson | c****e@a****m | 20 |
| Adam Tyson | a****n@u****k | 1 |
| Justin Kiggins | j****s@g****m | 1 |
| Simão Bolota | 9****l@u****m | 1 |
Committer Domains (Top 20 + Academic)
Packages
- Total packages: 2
-
Total downloads:
- pypi 93 last-month
-
Total dependent packages: 2
(may contain duplicates) -
Total dependent repositories: 0
(may contain duplicates) - Total versions: 17
- Total maintainers: 1
proxy.golang.org: github.com/brainglobe/brainreg-napari
- Documentation: https://pkg.go.dev/github.com/brainglobe/brainreg-napari#section-documentation
- License: mit
-
Latest release: v0.1.4
published about 2 years ago
Rankings
pypi.org: brainreg-napari
Multi-atlas whole-brain microscopy registration
- Documentation: https://brainreg-napari.readthedocs.io/
- License: BSD-3-Clause
-
Latest release: 0.1.4
published about 2 years ago
Rankings
Maintainers (1)
Dependencies
- actions/checkout v2 composite
- chanzuckerberg/napari-hub-preview-action v0.1 composite
- GabrielBB/xvfb-action v1 composite
- actions/checkout v3 composite
- actions/download-artifact v3 composite
- actions/setup-python v4 composite
- codecov/codecov-action v2 composite
- neuroinformatics-unit/actions/build_sdist_wheels main composite
- neuroinformatics-unit/actions/check_manifest v1.2.0 composite
- neuroinformatics-unit/actions/lint v1.2.0 composite
- pypa/gh-action-pypi-publish v1.5.0 composite
- tlambert03/setup-qt-libs v1 composite




