Science Score: 77.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
✓CITATION.cff file
Found CITATION.cff file -
✓codemeta.json file
Found codemeta.json file -
✓.zenodo.json file
Found .zenodo.json file -
✓DOI references
Found 4 DOI reference(s) in README -
✓Academic publication links
Links to: zenodo.org -
✓Committers with academic emails
3 of 5 committers (60.0%) from academic institutions -
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (16.2%) to scientific vocabulary
Keywords
Repository
Magnetic resonance imaging and tractography with R
Basic Info
Statistics
- Stars: 28
- Watchers: 5
- Forks: 8
- Open Issues: 1
- Releases: 10
Topics
Metadata Files
README.md
TractoR is an R-based platform for medical image processing, tractography and graph analysis. It is focussed on handling magnetic resonance images, but many of its core functions are applicable to any kind of image. The package is full-stack free and open-source, as R is itself free software, and it is easy to extend and build upon. Downloads are available from the project web site.
Overview
There are several sources of documentation available. For installation instructions, please see the INSTALL.md file. For general information on the package, its capabilities and conventions, please see the TractoR paper and/or the project home page. The latter is mirrored in the TractoR distribution, and can be found at share/doc/home.html. Standard R documentation is available, function by function, for the core tractor.base package and all first- and third-party dependency packages (which are included in the main download).
Please see below for information on
depending on your preference.
Usage from R
Users who are already familiar with R can easily incorporate the functionality of TractoR's R packages into their work. For example, an MRI image can be read from a compressed NIfTI-1 file, and its key metadata displayed with
```r t1 <- readImageFile("refT1.nii.gz") t1
Image source : /usr/local/tractor/tests/data/session/tractor/structural/refT1
Image dimensions : 176 x 232 x 256 voxels
Voxel dimensions : 1 x 1 x 1 mm
Coordinate origin : (91.89,94.98,106.1)
Additional tags : (none)
Sparseness : 10.03% (dense storage)
```
The t1 object can in many ways be treated as a standard R array, but it can also be manipulated and visualised with a set of specialised functions.
A parcellation in the same space can be loaded and overlaid in an interactive viewer using
r
parc <- readImageFile("parcellation.nii.gz")
showImagesInViewer(t1, parc)
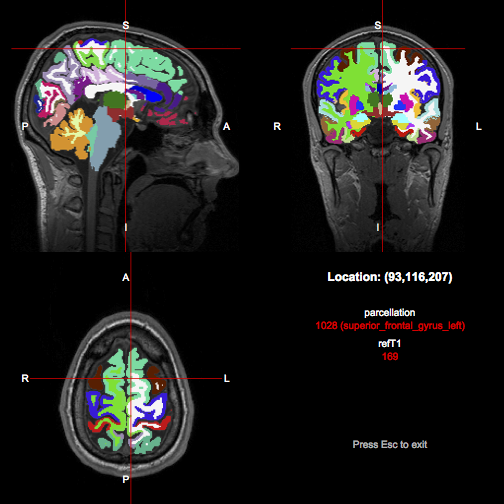
(with the tractor.session package loaded). Notice that both the numerical index and the name of the region currently under the crosshair is given in the info panel at the bottom right.
We can also apply image processing operations to the image and then visualise the result. In the example below we create a diamond-shaped 3D kernel, and then calculate and visualise the morphological gradient of the original image.
r
k <- shapeKernel(c(3,3,3), type="diamond")
grad <- t1$copy()$map(function(x) dilate(x,k) - erode(x,k))
showImagesInViewer(grad)
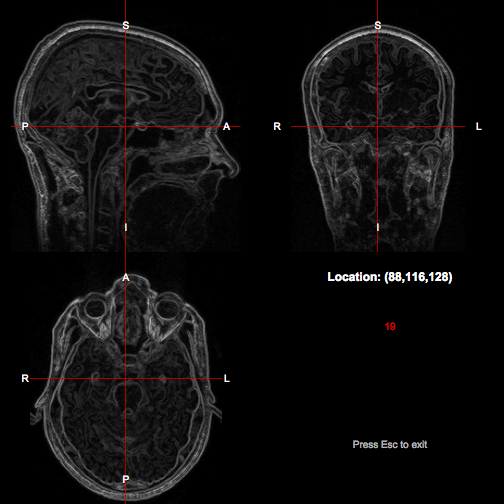
Notice the use of the map() method, which can be used to replace the data in an image with the result of applying a function, in this case an anonymous function constructed within the call.
Usage outside R
Users who are not familiar with R, or wish to perform common tasks quickly, can instead use TractoR's direct command-line interface, which is simpler to use and requires less typing, but is still very flexible. A list of available commands can be obtained with
```shell tractor list
Starting TractoR environment...
Experiment scripts found in /usr/local/tractor/share/experiments:
[1] age apply bedpost binarise
[5] chfiletype clone components compress
[9] console dicomread dicomsort dicomtags
[13] dirviz dpreproc extract freesurf
[17] gradcheck gradread gradrotate graph-build
[21] graph-decompose graph-extract graph-props graph-reweight
[25] graph-viz graph2csv hnt-eval hnt-interpret
[29] hnt-ref hnt-viz imageinfo imagestats
[33] import list mean mkroi
[37] morph parcellate path peek
[41] platform plotcorrections pnt-data pnt-em
[45] pnt-eval pnt-interpret pnt-prune pnt-ref
[49] pnt-train pnt-viz reg-apply reg-check
[53] reg-info reg-linear reg-nonlinear reg-viz
[57] reshape slice smooth split
[61] status tensorfit track transform
[65] trim update values view
Experiment completed with 0 warning(s) and 0 error(s)
```
Information on usage of a particular script, and the options that it supports, is available by using the '-o' command-line flag.
```shell tractor -o imagestats
OPTIONS for script /usr/local/tractor/share/experiments/imagestats.R (*
required)
Scope: nonzero [zero,positive,negative,all]
MaskFile: NULL
ThresholdLevel: 0
ARGUMENTS: image file
DESCRIPTION:
Print various statistics of the specified image, optionally limiting the
calculations by voxel value or by mask. The 5% trimmed range is obtained by
discarding the 5% largest and smallest values and taking the range of the
remainder, thereby giving an indication of the influence of extreme values.
This script may currently only be applied to 3D images.
```
Running a particular script is a matter of typing tractor, followed by the script's name and any arguments or named options required. For example,
shell
tractor graph-viz data/graph/diffusion ShowBrain:true
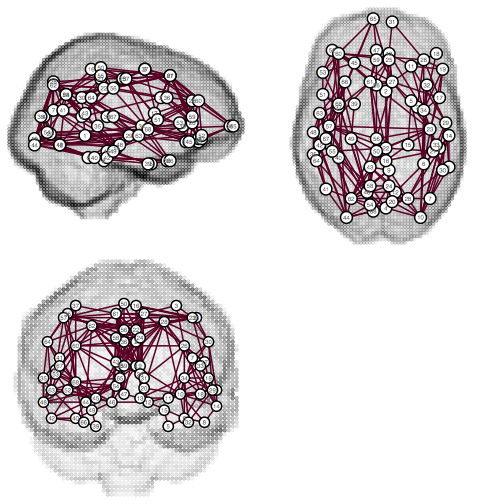
The examples in the previous section can be replicated in this way by using the imageinfo and view scripts. Try it! Further information on the tractor command is available from its man page, by typing man tractor.
Owner
- Name: TractoR
- Login: tractor
- Kind: organization
- Location: London, UK
- Website: www.tractor-mri.org.uk
- Repositories: 11
- Profile: https://github.com/tractor
The TractoR MRI analysis project and related code
Citation (CITATION.cff)
cff-version: 1.2.0
title: TractoR
message: >-
If you use this software, please cite it using the
metadata from this file.
type: software
authors:
- given-names: Jonathan D
family-names: Clayden
email: code@clayden.org
affiliation: University College London
orcid: 'https://orcid.org/0000-0002-6608-0619'
- given-names: Susana
family-names: Muñoz Maniega
affiliation: The University of Edinburgh
orcid: 'https://orcid.org/0000-0001-5185-6384'
- given-names: Fani
family-names: Deligianni
affiliation: University College London
orcid: 'https://orcid.org/0000-0003-1306-5017'
- given-names: Lena
family-names: Dorfschmidt
affiliation: University College London
- given-names: Kiran
family-names: Seunarine
affiliation: University College London
repository-code: 'https://github.com/tractor/tractor'
url: 'https://www.tractor-mri.org.uk'
keywords:
- MRI
- neuroimaging
- R
license: GPL-2.0
GitHub Events
Total
- Release event: 1
- Issue comment event: 1
- Push event: 72
- Create event: 3
Last Year
- Release event: 1
- Issue comment event: 1
- Push event: 72
- Create event: 3
Committers
Last synced: 9 months ago
Top Committers
| Name | Commits | |
|---|---|---|
| Jon Clayden | c****e@c****g | 3,101 |
| Fani Deligianni | f****i@u****k | 21 |
| Lena Dorfschmidt | l****t@u****k | 3 |
| Kiran Seunarine | k****e@u****k | 2 |
| John Muschelli | m****2@g****m | 1 |
Committer Domains (Top 20 + Academic)
Issues and Pull Requests
Last synced: 6 months ago
All Time
- Total issues: 6
- Total pull requests: 1
- Average time to close issues: 5 months
- Average time to close pull requests: about 1 hour
- Total issue authors: 5
- Total pull request authors: 1
- Average comments per issue: 4.67
- Average comments per pull request: 1.0
- Merged pull requests: 1
- Bot issues: 0
- Bot pull requests: 0
Past Year
- Issues: 0
- Pull requests: 0
- Average time to close issues: N/A
- Average time to close pull requests: N/A
- Issue authors: 0
- Pull request authors: 0
- Average comments per issue: 0
- Average comments per pull request: 0
- Merged pull requests: 0
- Bot issues: 0
- Bot pull requests: 0
Top Authors
Issue Authors
- jonclayden (2)
- muschellij2 (1)
- howff (1)
- resynth1943 (1)
- dominicjob (1)
Pull Request Authors
- muschellij2 (1)
Top Labels
Issue Labels
Pull Request Labels
Packages
- Total packages: 1
-
Total downloads:
- cran 740 last-month
- Total docker downloads: 43,464
- Total dependent packages: 2
- Total dependent repositories: 7
- Total versions: 31
- Total maintainers: 1
cran.r-project.org: tractor.base
Read, Manipulate and Visualise Magnetic Resonance Images
- Homepage: https://www.tractor-mri.org.uk
- Documentation: http://cran.r-project.org/web/packages/tractor.base/tractor.base.pdf
- License: GPL-2
-
Latest release: 3.4.2
published almost 2 years ago
Rankings
Maintainers (1)
Dependencies
- oro.nifti * enhances
- RNifti * imports
- methods * imports
- ore >= 1.3.0 imports
- reportr * imports
- shades * imports
- divest * suggests
- jsonlite * suggests
- loder * suggests
- mmand * suggests
- testthat * suggests
- yaml * suggests
- R >= 2.12.1 depends
- grDevices * depends
- graphics * depends
- methods * depends
- stats * depends
- utils * depends
- Rcpp * imports
- ore * imports
- reportr * imports
- tractor.base * imports
- boot * suggests
- igraph * suggests
- lattice * suggests
- testthat * suggests
- R >= 2.12.1 depends
- methods * depends
- ore * imports
- reportr * imports
- splines * imports
- tractor.base * imports
- tractor.session * imports
- tractor.track * imports
- tractor.reg * suggests
- RNiftyReg * imports
- methods * imports
- ore * imports
- reportr * imports
- tractor.base * imports
- RNifti * suggests
- R >= 2.12.1 depends
- methods * depends
- ore * imports
- reportr * imports
- tractor.base * imports
- yaml * imports
- RNiftyReg * suggests
- mmand * suggests
- tractor.reg * suggests
- tractor.track * suggests
- R >= 2.12.1 depends
- methods * depends
- RNifti >= 0.5.0 imports
- Rcpp * imports
- ore * imports
- reportr * imports
- tractor.base * imports
- tractor.reg * suggests
- R >= 2.12.1 depends
- utils * depends
- ore * imports
- reportr * imports
- tractor.base * imports
- parallel * suggests
- tractor.session * suggests
- yaml * suggests
- actions/cache v3 composite
- actions/checkout v3 composite
- r-lib/actions/setup-r v2 composite
- actions/checkout v3 composite
- rocker/r-base latest build