cytodataframe
An in-memory data analysis format for single-cell profiles alongside their corresponding images and segmentation masks.
Science Score: 77.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
✓CITATION.cff file
Found CITATION.cff file -
✓codemeta.json file
Found codemeta.json file -
✓.zenodo.json file
Found .zenodo.json file -
✓DOI references
Found 3 DOI reference(s) in README -
✓Academic publication links
Links to: zenodo.org -
✓Committers with academic emails
1 of 3 committers (33.3%) from academic institutions -
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (18.1%) to scientific vocabulary
Keywords
Keywords from Contributors
Repository
An in-memory data analysis format for single-cell profiles alongside their corresponding images and segmentation masks.
Basic Info
- Host: GitHub
- Owner: cytomining
- License: bsd-3-clause
- Language: Python
- Default Branch: main
- Homepage: https://cytomining.github.io/CytoDataFrame
- Size: 70.5 MB
Statistics
- Stars: 2
- Watchers: 1
- Forks: 2
- Open Issues: 23
- Releases: 14
Topics
Metadata Files
README.md

CytoDataFrame
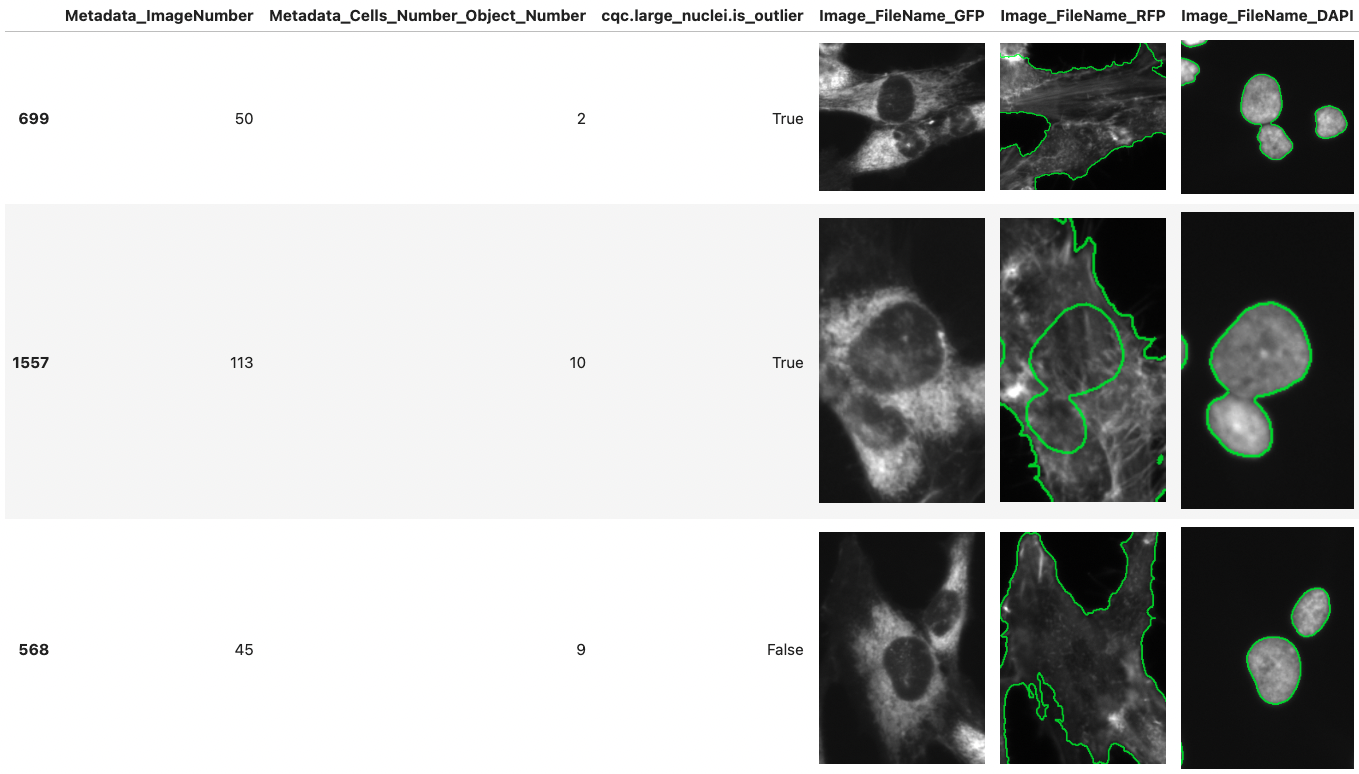 CytoDataFrame extends Pandas functionality to help display single-cell profile data alongside related images.
CytoDataFrame extends Pandas functionality to help display single-cell profile data alongside related images.
CytoDataFrame is an advanced in-memory data analysis format designed for single-cell profiling, integrating not only the data profiles but also their corresponding microscopy images and segmentation masks. Traditional single-cell profiling often excludes the associated images from analysis, limiting the scope of research. CytoDataFrame bridges this gap, offering a purpose-built solution for comprehensive analysis that incorporates both the data and images, empowering more detailed and visual insights in single-cell research.
CytoDataFrame is best suited for work within Jupyter notebooks. With CytoDataFrame you can:
- View image objects alongside their feature data using a Pandas DataFrame-like interface.
- Highlight image objects using mask or outline files to understand their segmentation.
- Adjust image displays on-the-fly using interactive slider widgets.
📓 Want to see CytoDataFrame in action? Check out our example notebook for a quick tour of its key features.
✨ CytoDataFrame development began within coSMicQC - a single-cell profile quality control package. Please check out our work there as well!
Installation
Install CytoDataFrame from source using the following:
```shell
install from pypi
pip install cytodataframe
or install directly from source
pip install git+https://github.com/cytomining/CytoDataFrame.git ```
Contributing, Development, and Testing
Please see our contributing documentation for more details on contributions, development, and testing.
References
Owner
- Name: cytomining
- Login: cytomining
- Kind: organization
- Repositories: 27
- Profile: https://github.com/cytomining
Citation (CITATION.cff)
# This CITATION.cff file was generated with cffinit.
# Visit https://bit.ly/cffinit to generate yours today!
---
cff-version: 1.2.0
title: CytoDataFrame
message: >-
If you use this software, please cite it using the
metadata from this file.
type: software
authors:
- given-names: David
family-names: Bunten
orcid: 'https://orcid.org/0000-0001-6041-3665'
- given-names: Jenna
family-names: Tomkinson
orcid: 'https://orcid.org/0000-0003-2676-5813'
- given-names: Vincent
family-names: Rubinetti
orcid: 'https://orcid.org/0000-0002-4655-3773'
- given-names: Gregory
family-names: Way
orcid: 'https://orcid.org/0000-0002-0503-9348'
repository-code: 'https://github.com/cytomining/CytoDataFrame'
identifiers:
- description: Software DOI
type: doi
value: "10.5281/zenodo.14797074"
abstract: >-
An in-memory data analysis format for single-cell profiles alongside their corresponding images and segmentation masks.
keywords:
- python
- single-cell-analysis
- profiling
- dataframes
- data-analysis
- way-lab
license: BSD-3-Clause
references:
- authors:
- name: "Way Lab CFReT_data Team"
date-accessed: "2024-05-13"
title: Way Lab CFReT_data CytoTable Data
type: data
repository-code: "https://github.com/WayScience/CFReT_data"
url: "https://github.com/WayScience/CFReT_data/blob/main/3.process_cfret_features/data/converted_profiles/localhost231120090001_converted.parquet"
scope: "localhost231120090001_converted.parquet"
notes: >-
Data from CFReT_data project is used to help validate
expected results. Data is generated from CellProfiler
and CytoTable.
identifiers:
- description: "Github Link with Contributors"
type: url
value: "https://github.com/WayScience/CFReT_data/graphs/contributors"
- authors:
- name: "Way Lab NF1_cellpainting_data Team"
date-accessed: "2024-06-28"
title: Way Lab NF1_cellpainting_data CytoTable Data
type: data
repository-code: "https://github.com/WayScience/nf1_cellpainting_data"
notes: >-
Data from NF1_cellpainting_data project is used to help validate
expected results. Data is generated from CellProfiler
and CytoTable. We use the following files from the repository:
- "Plate_2_nf1_analysis.sqlite"
- "Plate_2.parquet"
identifiers:
- description: "Github Link with Contributors"
type: url
value: "https://github.com/WayScience/nf1_cellpainting_data/graphs/contributors"
- title: >-
Plate 2 (Cell Painting images from Plate 2 for NF1_cellpainting_data project)
type: data
url: https://figshare.com/articles/dataset/Plate_2/22233700
notes: >-
Image data for related NF1_cellpainting_data parquet sqlite.
authors:
- family-names: Tomkinson
given-names: Jenna
- family-names: Mattson-Hoss
given-names: Michelle
- family-names: Sarnoff
given-names: Herb
- family-names: Way
given-names: Gregory
date-published: "2023-04-12"
identifiers:
- type: doi
value: 10.6084/m9.figshare.22233700.v4
- authors:
- name: "Way Lab and Alexander Lab Nuclear Speckles Collaboration"
date-accessed: "2024-09-04"
title: Way Lab and Alexander Lab Nuclear Speckles Collaboration Data
type: data
repository-code: https://github.com/WayScience/nuclear_speckle_image_profiling
notes: >-
Data from a collaborative project focusing on nuclear speckles
with the Way Lab and Alexander Lab s used to help validate
expected results. Parquet data is generated from CellProfiler
and CytoTable. Images courtesy of Katherine Alexander
and the Alexander Lab.
identifiers:
- description: "Github Link with Contributors"
type: url
value: "https://github.com/WayScience/nuclear_speckle_image_profiling/graphs/contributors"
- authors:
- family-names: Chandrasekaran
given-names: Srinivas Niranj
- family-names: Cimini
given-names: Beth A.
- family-names: Goodale
given-names: Amy
- family-names: Miller
given-names: Lisa
- family-names: Kost-Alimova
given-names: Maria
- family-names: Jamali
given-names: Nasim
- family-names: Doench
given-names: John G.
- family-names: Fritchman
given-names: Briana
- family-names: Skepner
given-names: Adam
- family-names: Melanson
given-names: Michelle
- family-names: Kalinin
given-names: Alexandr A.
- family-names: Arevalo
given-names: John
- family-names: Haghighi
given-names: Marzieh
- family-names: Caicedo
given-names: Juan C.
- family-names: Kuhn
given-names: Daniel
- family-names: Hernandez
given-names: Desiree
- family-names: Berstler
given-names: James
- family-names: Shafqat-Abbasi
given-names: Hamdah
- family-names: Root
given-names: David E.
- family-names: Swalley
given-names: Susanne E.
- family-names: Garg
given-names: Sakshi
- family-names: Singh
given-names: Shantanu
- family-names: Carpenter
given-names: Anne E.
date-accessed: "2024-08-21"
title: >-
Three million images and morphological profiles of cells treated with matched chemical and genetic perturbations
type: article
issn: 1548-7105
issue: 6
journal: Nature Methods
pages: 1114-1121
volume: 21
url: https://doi.org/10.1038/s41592-024-02241-6
date-published: "2024-06-01"
identifiers:
- type: doi
value: 10.1038/s41592-024-02241-6
notes: >-
JUMP (cpg0000-jump-pilot) was used to help demonstrate CytoDataFrame performance
with large data. See here for more information:
https://github.com/broadinstitute/cellpainting-gallery
GitHub Events
Total
- Create event: 3
- Release event: 1
- Issues event: 9
- Watch event: 2
- Delete event: 2
- Issue comment event: 11
- Push event: 21
- Pull request review comment event: 28
- Pull request review event: 31
- Pull request event: 17
- Fork event: 1
Last Year
- Create event: 3
- Release event: 1
- Issues event: 9
- Watch event: 2
- Delete event: 2
- Issue comment event: 11
- Push event: 21
- Pull request review comment event: 28
- Pull request review event: 31
- Pull request event: 17
- Fork event: 1
Committers
Last synced: 9 months ago
Top Committers
| Name | Commits | |
|---|---|---|
| dependabot[bot] | 4****] | 39 |
| Dave Bunten | d****n@c****u | 25 |
| Jenna Tomkinson | 1****n | 1 |
Committer Domains (Top 20 + Academic)
Issues and Pull Requests
Last synced: 9 months ago
All Time
- Total issues: 32
- Total pull requests: 68
- Average time to close issues: 24 days
- Average time to close pull requests: 2 days
- Total issue authors: 2
- Total pull request authors: 3
- Average comments per issue: 0.44
- Average comments per pull request: 0.72
- Merged pull requests: 60
- Bot issues: 0
- Bot pull requests: 41
Past Year
- Issues: 31
- Pull requests: 68
- Average time to close issues: 24 days
- Average time to close pull requests: 2 days
- Issue authors: 2
- Pull request authors: 3
- Average comments per issue: 0.39
- Average comments per pull request: 0.72
- Merged pull requests: 60
- Bot issues: 0
- Bot pull requests: 41
Top Authors
Issue Authors
- d33bs (33)
- jenna-tomkinson (5)
Pull Request Authors
- dependabot[bot] (40)
- d33bs (35)
- jenna-tomkinson (1)
- vincerubinetti (1)
Top Labels
Issue Labels
Pull Request Labels
Dependencies
- release-drafter/release-drafter v6 composite
- JamesIves/github-pages-deploy-action v4 composite
- actions/checkout v4 composite
- actions/setup-python v5 composite
- actions/checkout v4 composite
- actions/setup-python v5 composite
- pypa/gh-action-pypi-publish release/v1 composite
- actions/checkout v4 composite
- actions/setup-python v5 composite
- pre-commit/action v3.0.1 composite
- 110 dependencies



