brainreg
Automated 3D brain registration with support for multiple species and atlases.
Science Score: 49.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
○CITATION.cff file
-
✓codemeta.json file
Found codemeta.json file -
✓.zenodo.json file
Found .zenodo.json file -
✓DOI references
Found 8 DOI reference(s) in README -
✓Academic publication links
Links to: nature.com -
○Academic email domains
-
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (13.8%) to scientific vocabulary
Keywords
Repository
Automated 3D brain registration with support for multiple species and atlases.
Basic Info
- Host: GitHub
- Owner: brainglobe
- License: bsd-3-clause
- Language: Python
- Default Branch: main
- Homepage: https://brainglobe.info/documentation/brainreg/index.html
- Size: 821 MB
Statistics
- Stars: 136
- Watchers: 8
- Forks: 32
- Open Issues: 19
- Releases: 16
Topics
Metadata Files
README.md
brainreg
brainreg is an update to amap (which is itself a port of the original Java software) to include multiple registration backends, and to support the many atlases provided by brainglobe-atlasapi. It also comes with an optional napari plugin if you'd rather use brainreg with through graphical interface.
Documentation for both the command-line tool and graphical interface can be found here.
For segmentation of bulk structures in 3D space (e.g. injection sites, Neuropixels probes), please see brainglobe-segmentation.
Details
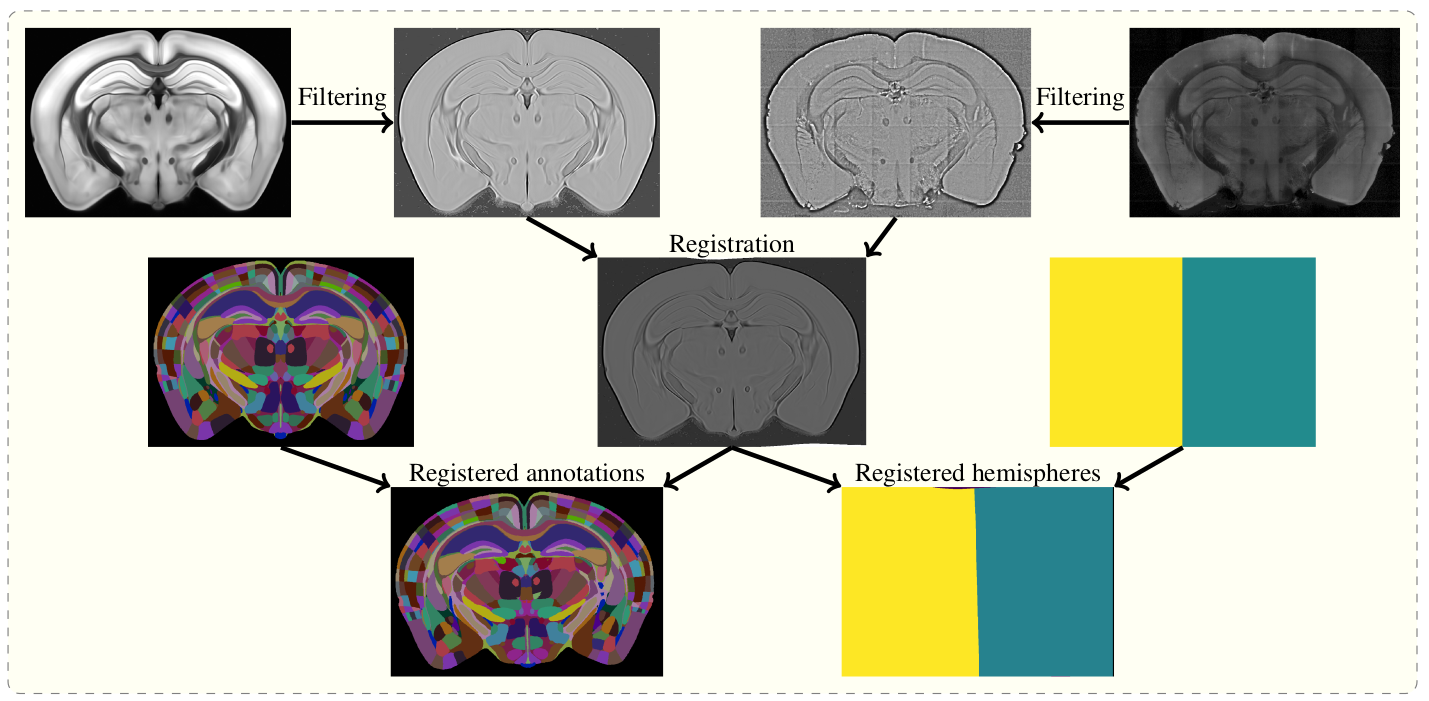
The aim of brainreg is to register the template brain (e.g. from the Allen Reference Atlas) to the sample image. Once this is complete, any other image in the template space can be aligned with the sample (such as region annotations, for segmentation of the sample image). The template to sample transformation can also be inverted, allowing sample images to be aligned in a common coordinate space.
To do this, the template and sample images are filtered, and then registered in a three step process (reorientation, affine registration, and freeform registration). The resulting transform from template to standard space is then applied to the atlas.
Full details of the process are in the original aMAP paper.
Installation
To install both the command line tool and the napari plugin, run
bash
pip install brainreg[napari]
in your desired Python environment. To only install the command line tool with no GUI (e.g. to run brainreg on an HPC cluster), just run:
bash
pip install brainreg
Installing on macOS
If you are using macOS, please run
bash
conda install -c conda-forge niftyreg
in your environment before installing, to ensure all dependencies are installed.
Command line usage
Basic usage
bash
brainreg /path/to/raw/data /path/to/output/directory -v 5 2 2 --orientation psl
Full command-line arguments are available with brainreg -h, but please
get in touch if you have any questions.
Mandatory arguments
- Path to the directory of the images. This can also be a text file pointing to the files.
- Output directory for all intermediate and final results.
- You must also specify the voxel sizes with the
-vflag, see specifying voxel size for details.
Atlas
By default, brainreg will use the 25um version of the Allen Mouse Brain Atlas.
To use another atlas (e.g. for another species, or another resolution), you must use the --atlas flag, followed by the string describing the atlas, e.g.:
bash
--atlas allen_mouse_50um
To find out which atlases are available, once brainreg is installed, please run brainglobe list.
The name of the resulting atlases is the string to pass with the --atlas flag.
Input data orientation
If your data does not match the BrainGlobe default orientation (the origin voxel is the most anterior, superior, left-most voxel), then you must specify the orientation by using the --orientation flag.
What follows must be a string in the brainglobe-space "initials" form, to describe the origin voxel.
If the origin of your data (first, top left voxel) is the most anterior, superior, left part of the brain, then the orientation string would be "asl" (anterior, superior, left), and you would use:
bash
--orientation asl
Registration options
To change how the actual registration performs, see registration parameters
Additional options
-aor--additionalPaths to N additional channels to downsample to the same coordinate space.--sort-input-fileIf set to true, the input text file will be sorted using natural sorting. This means that the file paths will be sorted as would be expected by a human and not purely alphabetically.--brain_geometryCan be one offull(default) for full brain registration,hemisphere_lfor left hemisphere data-set andhemisphere_rfor right hemisphere data-set.
Misc options
--n-free-cpusThe number of CPU cores on the machine to leave unused by the program to spare resources.--debugDebug mode. Will increase verbosity of logging and save all intermediate files for diagnosis of software issues.--save-original-orientationOption to save the registered atlas with the same orientation as the input data.
Visualising results
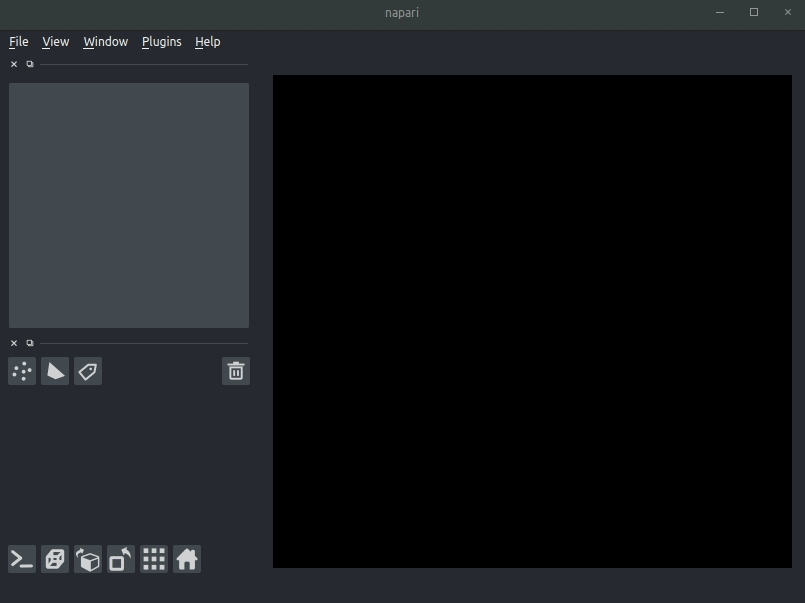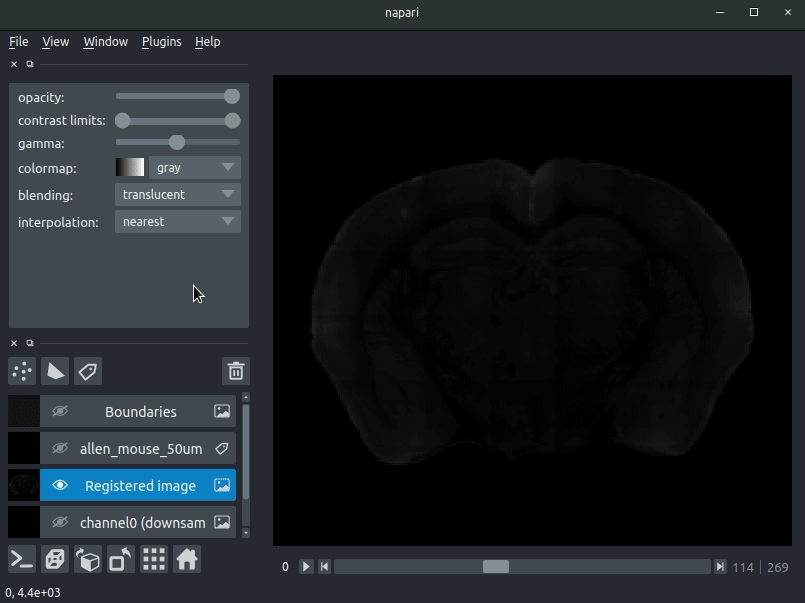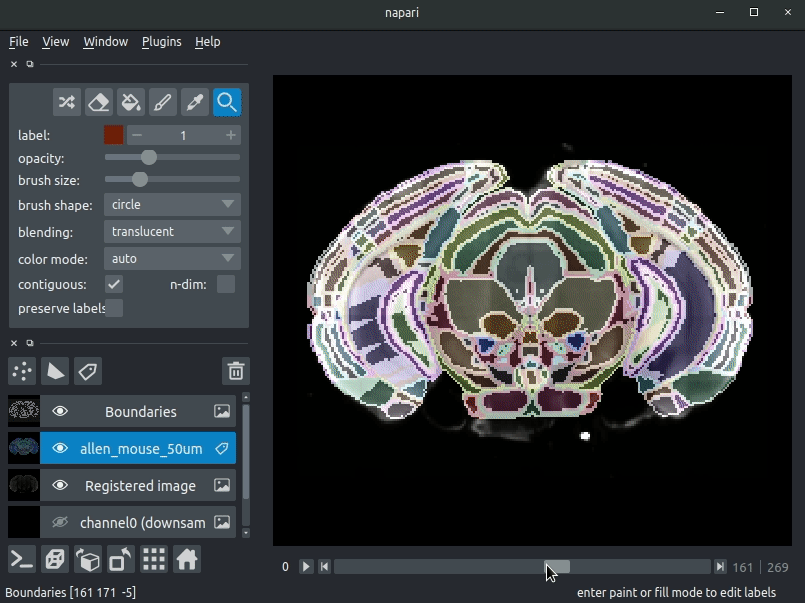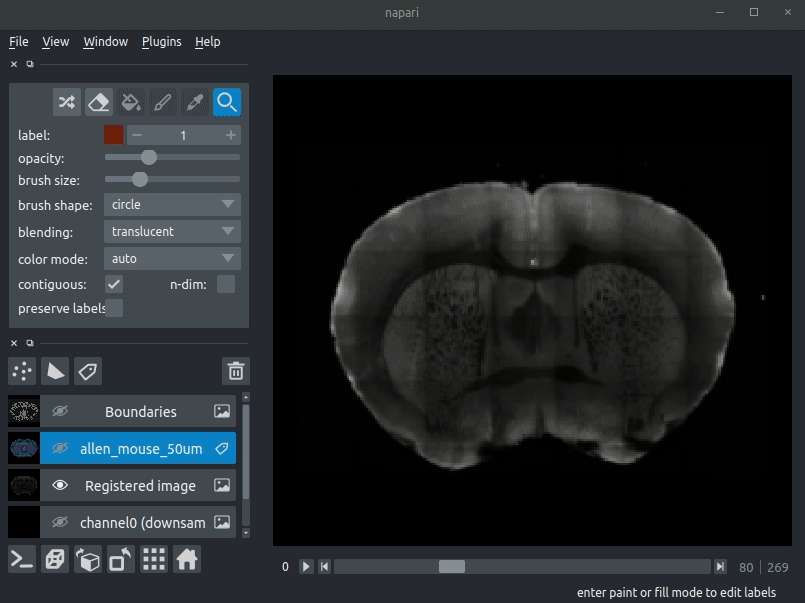
If you have installed the optional napari plugin, you can use napari to view your data. The plugin automatically fetches the brainglobe-napari-io which provides this functionality. If you have installed only the command-line tool you can still manually install brainglobe-napari-io and follow the steps below.
Sample space
Open napari and drag your brainreg output directory (the one with the log file) onto the napari window.
Various images should then open, including:
Registered image- the image used for registration, downsampled to atlas resolutionatlas_name- e.g.allen_mouse_25umthe atlas labels, warped to your sample brainBoundaries- the boundaries of the atlas regions
If you downsampled additional channels, these will also be loaded. Most of these images will not be visible by default - click the little eye icon to toggle visibility.
Note: If you use a high resolution atlas (such as allen_mouse_10um), then the files can take a little while to load.
Seeking help or contributing
We are always happy to help users of our tools, and welcome any contributions. If you would like to get in contact with us for any reason, please see the contact page of our website.
Citing brainreg
If you find brainreg useful, and use it in your research, please let us know and also cite the paper:
Tyson, A. L., Vélez-Fort, M., Rousseau, C. V., Cossell, L., Tsitoura, C., Lenzi, S. C., Obenhaus, H. A., Claudi, F., Branco, T., Margrie, T. W. (2022). Accurate determination of marker location within whole-brain microscopy images. Scientific Reports, 12, 867 doi.org/10.1038/s41598-021-04676-9
Please also cite aMAP (the original pipeline from which this software is based):
Niedworok, C.J., Brown, A.P.Y., Jorge Cardoso, M., Osten, P., Ourselin, S., Modat, M. and Margrie, T.W., (2016). AMAP is a validated pipeline for registration and segmentation of high-resolution mouse brain data. Nature Communications. 7, 19. https://doi.org/10.1038/ncomms11879
Lastly, if you can, please cite the BrainGlobe Atlas API that provided the atlas:
Claudi, F., Petrucco, L., Tyson, A. L., Branco, T., Margrie, T. W. and Portugues, R. (2020). BrainGlobe Atlas API: a common interface for neuroanatomical atlases. Journal of Open Source Software, 5(54), 2668, https://doi.org/10.21105/joss.02668
Finally, don't forget to cite the developers of the atlas that you used (e.g. the Allen Brain Atlas)!
Owner
- Name: BrainGlobe
- Login: brainglobe
- Kind: organization
- Location: London/Munich
- Website: https://brainglobe.info/
- Twitter: brain_globe
- Repositories: 28
- Profile: https://github.com/brainglobe
Open python tools for morphological analyses in systems neuroscience
GitHub Events
Total
- Create event: 18
- Release event: 2
- Issues event: 15
- Watch event: 15
- Delete event: 17
- Issue comment event: 32
- Push event: 28
- Pull request review event: 23
- Pull request event: 35
- Fork event: 7
Last Year
- Create event: 18
- Release event: 2
- Issues event: 15
- Watch event: 15
- Delete event: 17
- Issue comment event: 32
- Push event: 28
- Pull request review event: 23
- Pull request event: 35
- Fork event: 7
Issues and Pull Requests
Last synced: 6 months ago
All Time
- Total issues: 6
- Total pull requests: 18
- Average time to close issues: about 1 month
- Average time to close pull requests: about 11 hours
- Total issue authors: 6
- Total pull request authors: 7
- Average comments per issue: 2.17
- Average comments per pull request: 0.06
- Merged pull requests: 13
- Bot issues: 0
- Bot pull requests: 6
Past Year
- Issues: 5
- Pull requests: 18
- Average time to close issues: about 1 month
- Average time to close pull requests: about 11 hours
- Issue authors: 5
- Pull request authors: 7
- Average comments per issue: 1.4
- Average comments per pull request: 0.06
- Merged pull requests: 13
- Bot issues: 0
- Bot pull requests: 6
Top Authors
Issue Authors
- alessandrofelder (6)
- adamltyson (4)
- ClementCaporal (1)
- Yanting-Yao (1)
- yiyangcrick (1)
- AdrianAriasAbreu (1)
- samclothier (1)
- pderdeyn (1)
- dstansby (1)
- sama-maturi (1)
- willGraham01 (1)
- jingjie-li (1)
- Eddymorphling (1)
Pull Request Authors
- pre-commit-ci[bot] (17)
- adamltyson (14)
- alessandrofelder (8)
- IgorTatarnikov (8)
- willGraham01 (5)
- K-Meech (2)
- lochhh (1)
- thisisrick25 (1)
- Eddymorphling (1)
- healthonrails (1)
Top Labels
Issue Labels
Pull Request Labels
Packages
- Total packages: 1
-
Total downloads:
- pypi 1,379 last-month
- Total dependent packages: 6
- Total dependent repositories: 1
- Total versions: 49
- Total maintainers: 1
pypi.org: brainreg
Automated multi-atlas whole-brain microscopy registration
- Homepage: https://brainglobe.info
- Documentation: https://docs.brainglobe.info/brainreg
- License: BSD 3-Clause License Copyright (c) 2020, University College London All rights reserved. Redistribution and use in source and binary forms, with or without modification, are permitted provided that the following conditions are met: * Redistributions of source code must retain the above copyright notice, this list of conditions and the following disclaimer. * Redistributions in binary form must reproduce the above copyright notice, this list of conditions and the following disclaimer in the documentation and/or other materials provided with the distribution. * Neither the name of the copyright holder nor the names of its contributors may be used to endorse or promote products derived from this software without specific prior written permission. THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS "AS IS" AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE DISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT HOLDER OR CONTRIBUTORS BE LIABLE FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY, OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE OF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.
-
Latest release: 1.0.13
published 9 months ago
Rankings
Maintainers (1)
Dependencies
- actions/cache v3 composite
- actions/checkout v2 composite
- actions/setup-python v1 composite
- actions/setup-python v2 composite
- brainglobe/actions/lint v1 composite
- brainglobe/actions/test v1 composite
- bg-atlasapi *
- bg-space *
- brainglobe-utils *
- brainglobe-utils >=0.2.7
- fancylog *
- imio *
- numpy *
- scikit-image *





