karyoploteR
karyoploteR - An R/Bioconductor package to plot arbitrary data along the genome
Science Score: 49.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
○CITATION.cff file
-
✓codemeta.json file
Found codemeta.json file -
✓.zenodo.json file
Found .zenodo.json file -
✓DOI references
Found 3 DOI reference(s) in README -
○Academic publication links
-
✓Committers with academic emails
2 of 12 committers (16.7%) from academic institutions -
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (14.1%) to scientific vocabulary
Keywords
Keywords from Contributors
Repository
karyoploteR - An R/Bioconductor package to plot arbitrary data along the genome
Basic Info
- Host: GitHub
- Owner: bernatgel
- Language: R
- Default Branch: devel
- Homepage: https://bernatgel.github.io/karyoploter_tutorial/
- Size: 3.08 MB
Statistics
- Stars: 337
- Watchers: 6
- Forks: 44
- Open Issues: 66
- Releases: 2
Topics
Metadata Files
README.md
karyoploteR - An R/Biocondutor package to plot arbitrary data along the genome
Description
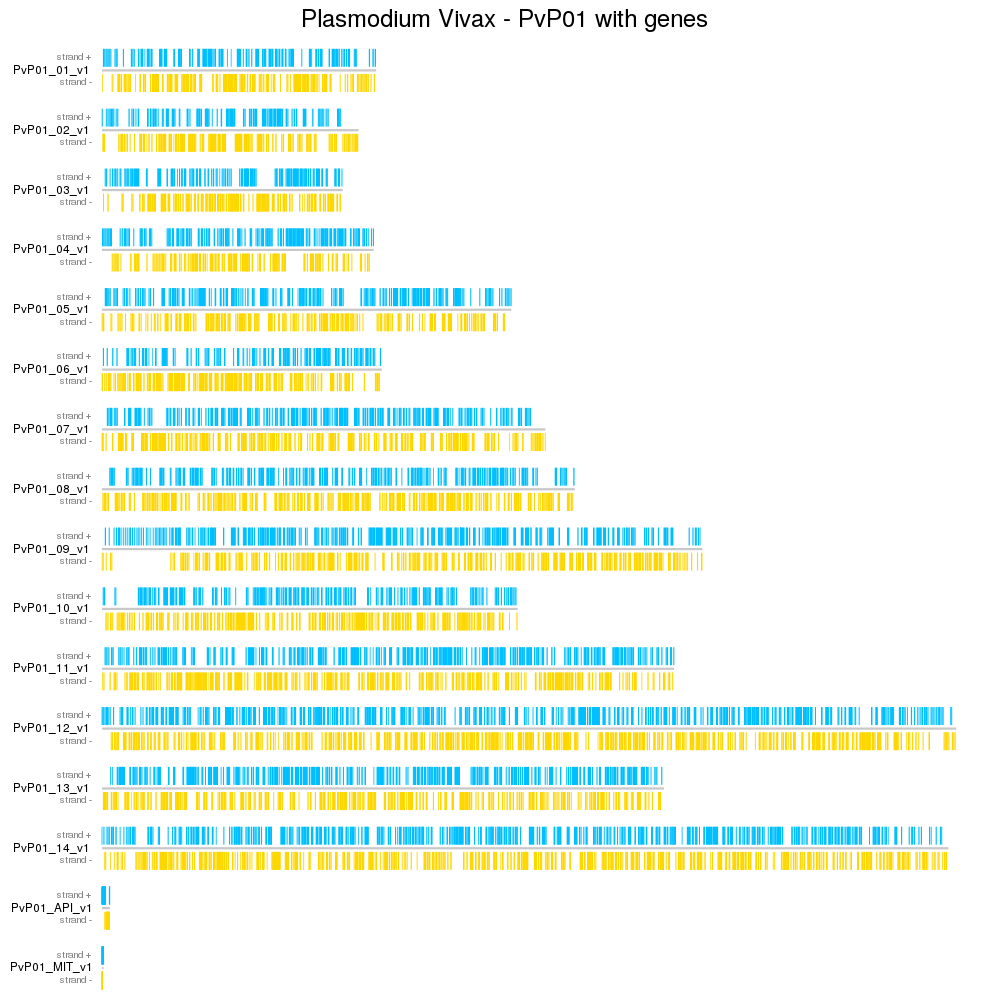
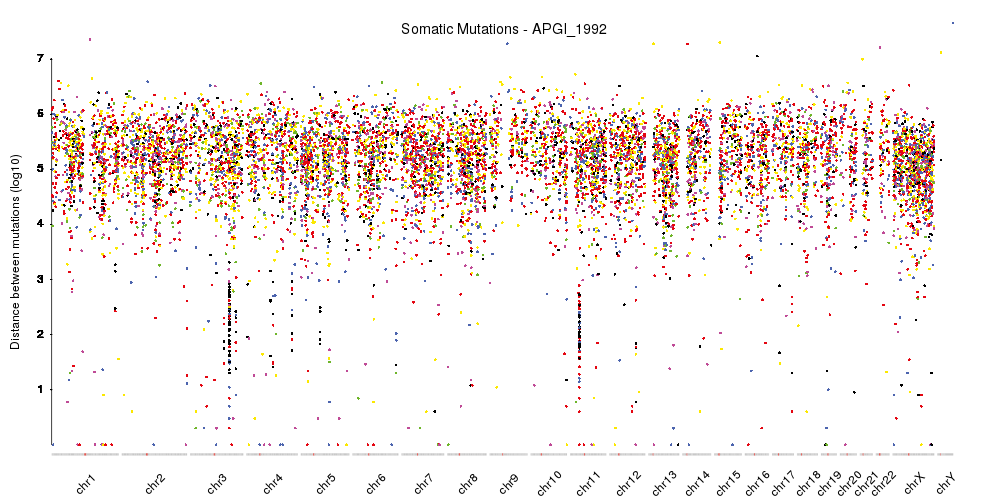
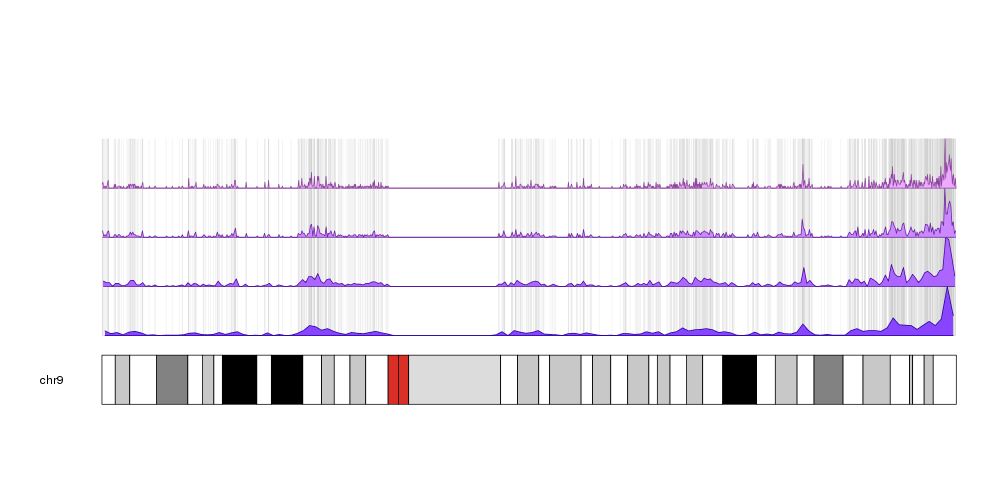
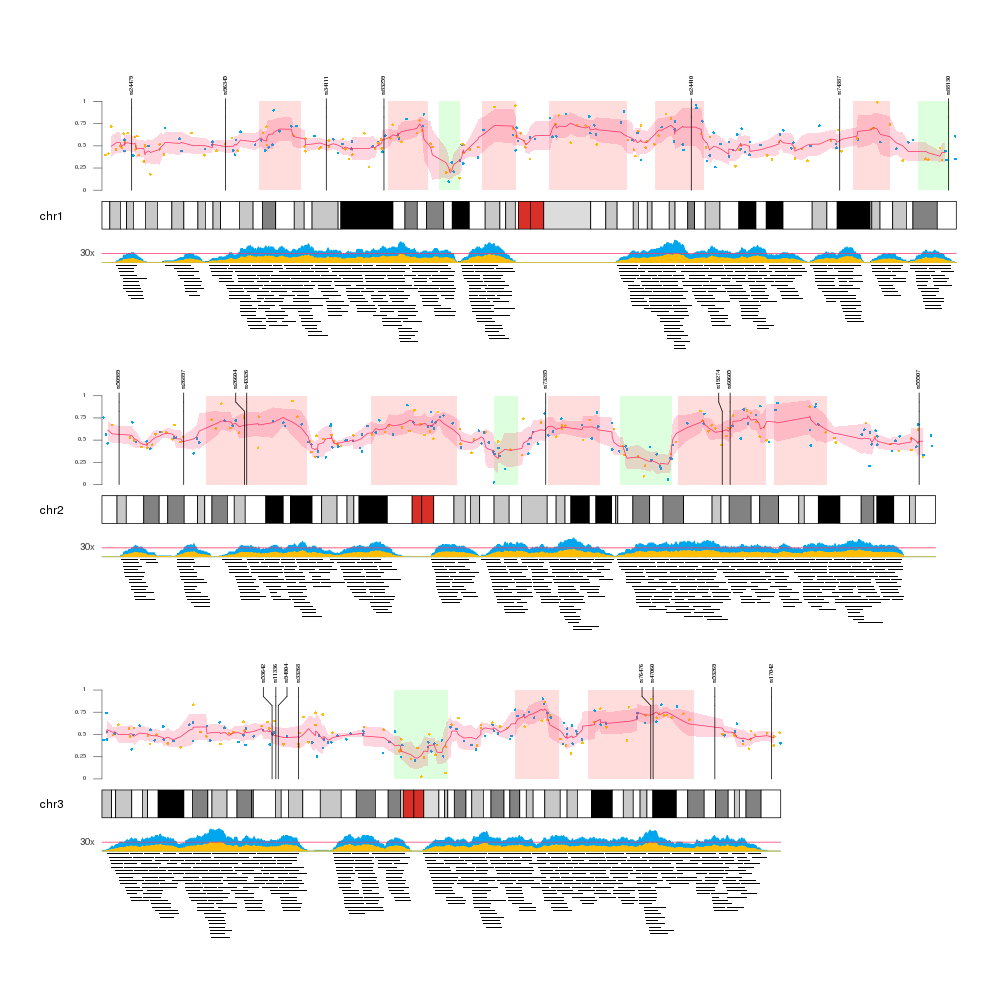
karyoploteR is an R package to plot data along the genome using a karyotype style plot.
It is entirely based on R base graphics and inspired by the R base graphics API. It includes functions to plot primitive graphic elements such as points, lines, rectangles, text, etc mapped into the genome plot coordinates and higher level functions to plot a heatmap, the regions in a GenomicRanges object or the cumulative coverage of such regions.
Data positioning and track configuration has been inspired by Circos and does not explicitly understands the concept of track. Thus, it is possible to freely specify where to plot the data and to create plots with multiple independent tracks or overlapping representations.
It is highly configurable and in addition to the parametrization of the different data plotting functions, it is possible to specify custom functions for every plotting action from the basic chromosome bands to the chromosome labels or base numbers as well as creating completely new plotting functions.
How to use it
Documentation (vignette and user manual) is available at the karyoploteR's Bioconductor landing page at http://bioconductor.org/packages/karyoploteR
Tutorial and Examples
In addition to the documentation above, a short tutorial and some examples can be found at https://bernatgel.github.io/karyoploter_tutorial/
Citing karyoploteR
karyoploteR has been developed by Bernat Gel and Eduard Serra at IGTP Hereditary Cancer Group.
If you use karyoploteR in your research, please cite the Bioinformatics paper describing it:
Bernat Gel & Eduard Serra. (2017). karyoploteR: an R/Bioconductor package to plot customizable genomes displaying arbitrary data. Bioinformatics, 31–33. doi:10.1093/bioinformatics/btx346
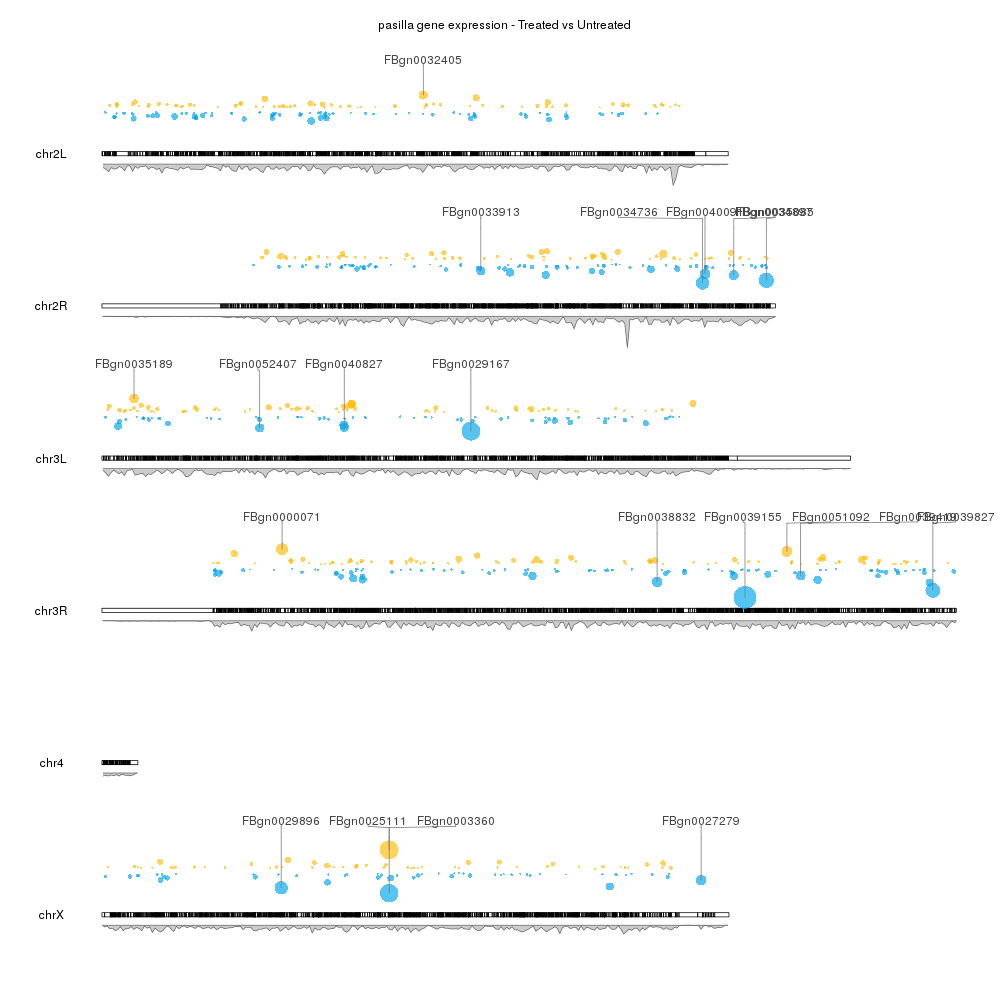
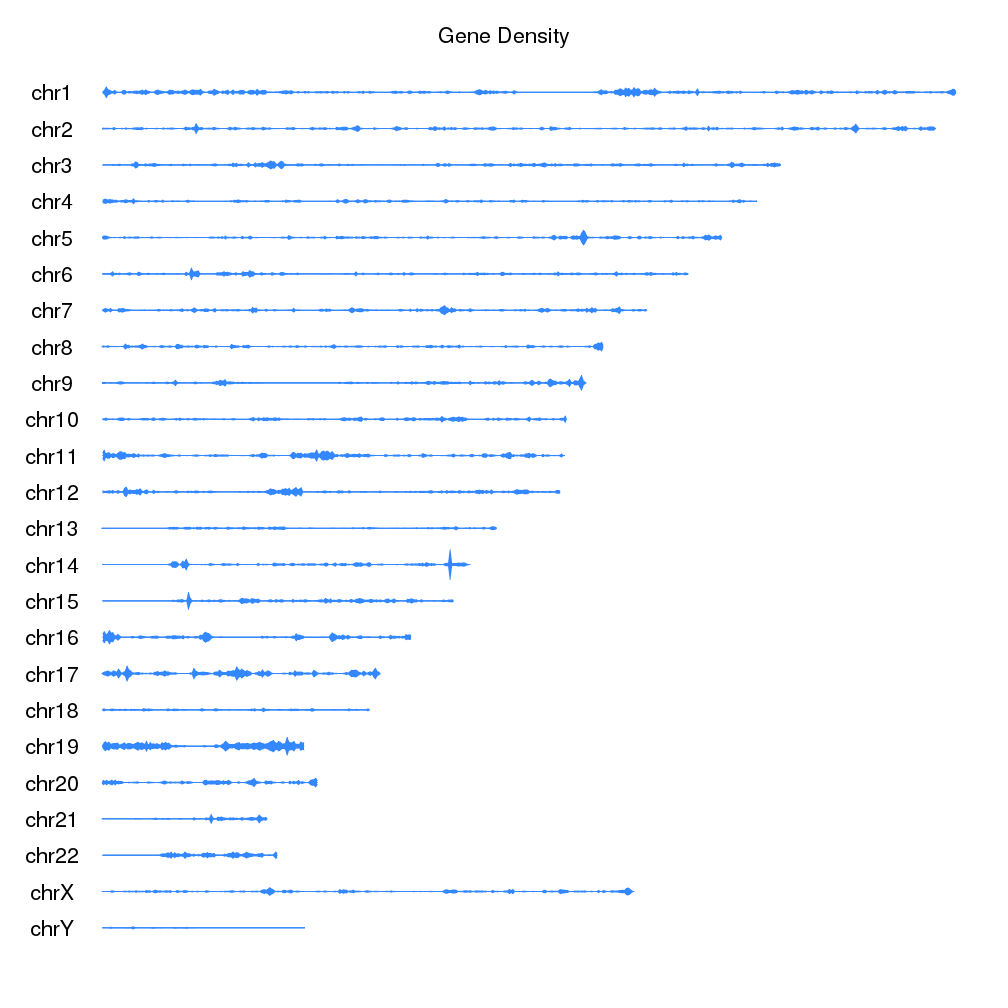
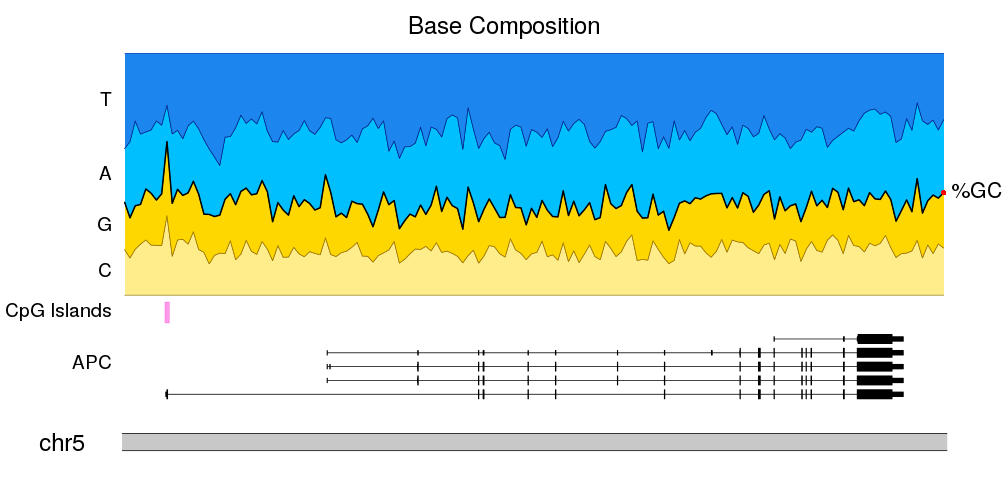
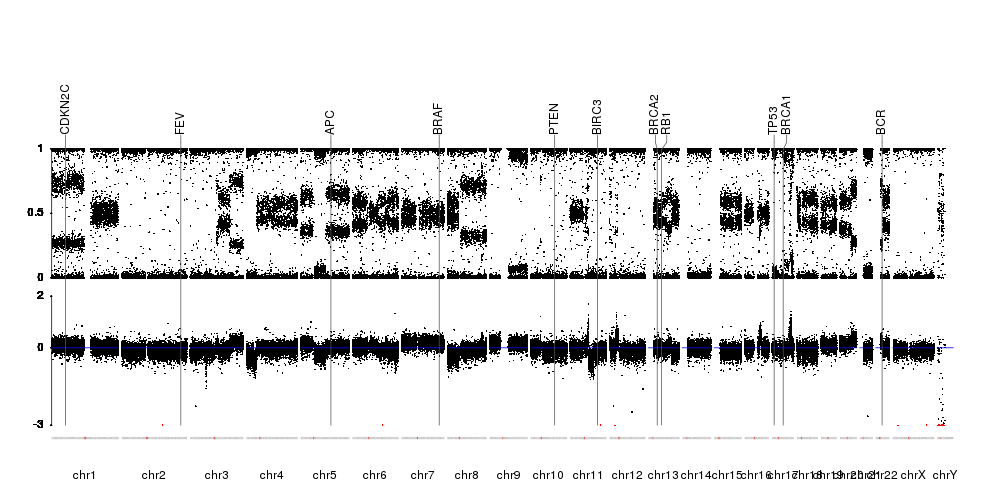
A few example plots created with karyoploteR
These images are all created with karyoploteR and are part of the documented examples in the karyoploteR's tutorial and examples page. Click on them to see how the code needed to create them.
Owner
- Name: Bernat Gel
- Login: bernatgel
- Kind: user
- Location: Barcelona, Spain
- Company: Germans Trias Research Institute (IGTP)
- Twitter: bernatgel
- Repositories: 4
- Profile: https://github.com/bernatgel
Bioinformatician at IGTP. Interested in cancer genomics, data analysis and visualization. Working on NF1, Hereditary Cancer and genetic diagnostics.
GitHub Events
Total
- Issues event: 8
- Watch event: 32
- Issue comment event: 4
- Push event: 2
- Fork event: 1
Last Year
- Issues event: 8
- Watch event: 32
- Issue comment event: 4
- Push event: 2
- Fork event: 1
Committers
Last synced: 7 months ago
Top Committers
| Name | Commits | |
|---|---|---|
| Bernat Gel | b****l@g****m | 398 |
| Bernat Gel | b****l@i****g | 128 |
| Nitesh Turaga | n****a@g****m | 14 |
| J Wokaty | j****y@s****u | 10 |
| Hervé Pagès | h****s@f****g | 3 |
| Bernat | b****l@g****m | 3 |
| vobencha | v****a@g****m | 2 |
| vobencha | v****n@r****g | 2 |
| Herve Pages | h****s@f****g | 2 |
| A Wokaty | a****y@s****u | 2 |
| Darío Hereñú | m****a@g****m | 1 |
| Martin Morgan | m****n@f****g | 1 |
Committer Domains (Top 20 + Academic)
Issues and Pull Requests
Last synced: 6 months ago
All Time
- Total issues: 118
- Total pull requests: 0
- Average time to close issues: 4 months
- Average time to close pull requests: N/A
- Total issue authors: 90
- Total pull request authors: 0
- Average comments per issue: 2.1
- Average comments per pull request: 0
- Merged pull requests: 0
- Bot issues: 0
- Bot pull requests: 0
Past Year
- Issues: 9
- Pull requests: 0
- Average time to close issues: N/A
- Average time to close pull requests: N/A
- Issue authors: 9
- Pull request authors: 0
- Average comments per issue: 0.89
- Average comments per pull request: 0
- Merged pull requests: 0
- Bot issues: 0
- Bot pull requests: 0
Top Authors
Issue Authors
- bernatgel (10)
- vdejager (4)
- lbeltrame (3)
- diyasen2021 (3)
- rthapa1 (3)
- yxian9 (2)
- lakhujanivijay (2)
- njbowen (2)
- jromanowska (2)
- gbdias (2)
- nirvana693 (2)
- zhoudreames (2)
- Mailinnia (2)
- TheSallyGardens (2)
- Hanuphant (2)
Pull Request Authors
Top Labels
Issue Labels
Pull Request Labels
Packages
- Total packages: 1
-
Total downloads:
- bioconductor 107,792 total
- Total dependent packages: 8
- Total dependent repositories: 0
- Total versions: 6
- Total maintainers: 1
bioconductor.org: karyoploteR
Plot customizable linear genomes displaying arbitrary data
- Homepage: https://github.com/bernatgel/karyoploteR
- Documentation: https://bioconductor.org/packages/release/bioc/vignettes/karyoploteR/inst/doc/karyoploteR.pdf
- License: Artistic-2.0
-
Latest release: 1.34.2
published 8 months ago
Rankings
Maintainers (1)
Dependencies
- GenomicRanges * depends
- R >= 3.4 depends
- methods * depends
- regioneR * depends
- AnnotationDbi * imports
- GenomeInfoDb * imports
- GenomicFeatures * imports
- GenomicRanges * imports
- IRanges * imports
- Rsamtools * imports
- S4Vectors * imports
- VariantAnnotation * imports
- bamsignals * imports
- bezier * imports
- biovizBase * imports
- digest * imports
- grDevices * imports
- graphics * imports
- memoise * imports
- regioneR * imports
- rtracklayer * imports
- stats * imports
- BSgenome.Hsapiens.UCSC.hg19 * suggests
- BSgenome.Hsapiens.UCSC.hg19.masked * suggests
- BiocStyle * suggests
- TxDb.Hsapiens.UCSC.hg19.knownGene * suggests
- TxDb.Mmusculus.UCSC.mm10.knownGene * suggests
- knitr * suggests
- magrittr * suggests
- markdown * suggests
- org.Hs.eg.db * suggests
- org.Mm.eg.db * suggests
- pasillaBamSubset * suggests
- rmarkdown * suggests
- testthat * suggests