icetea
R/Bioconductor package for analysis of single and paired-end capped-RNAseq data (CAGE/RAMPAGE/MAPCap)
Science Score: 20.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
○CITATION.cff file
-
○codemeta.json file
-
○.zenodo.json file
-
○DOI references
-
✓Academic publication links
Links to: nature.com -
✓Committers with academic emails
1 of 6 committers (16.7%) from academic institutions -
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (13.3%) to scientific vocabulary
Keywords
cage
expression
rna-seq
transcriptomics
Keywords from Contributors
bioconductor-package
gene
proteomics
genomics
expression-analysis
mendelian-genetics
outlier-detection
rpackage
dimension-reduction
feature-extraction
Last synced: 6 months ago
·
JSON representation
Repository
R/Bioconductor package for analysis of single and paired-end capped-RNAseq data (CAGE/RAMPAGE/MAPCap)
Basic Info
- Host: GitHub
- Owner: vivekbhr
- License: gpl-3.0
- Language: R
- Default Branch: master
- Homepage: https://vivekbhr.github.io/icetea/
- Size: 8.83 MB
Statistics
- Stars: 2
- Watchers: 0
- Forks: 0
- Open Issues: 3
- Releases: 0
Topics
cage
expression
rna-seq
transcriptomics
Created about 10 years ago
· Last pushed over 5 years ago
https://github.com/vivekbhr/icetea/blob/master/
# icetea
[](https://travis-ci.org/vivekbhr/icetea)
[](https://ci.appveyor.com/project/vivekbhr/icetea/branch/master)
[](https://coveralls.io/github/vivekbhr/icetea?branch=master)
*An R package for analysis of data produced by transcript 5' profiling methods like RAMPAGE and MAPCap.*
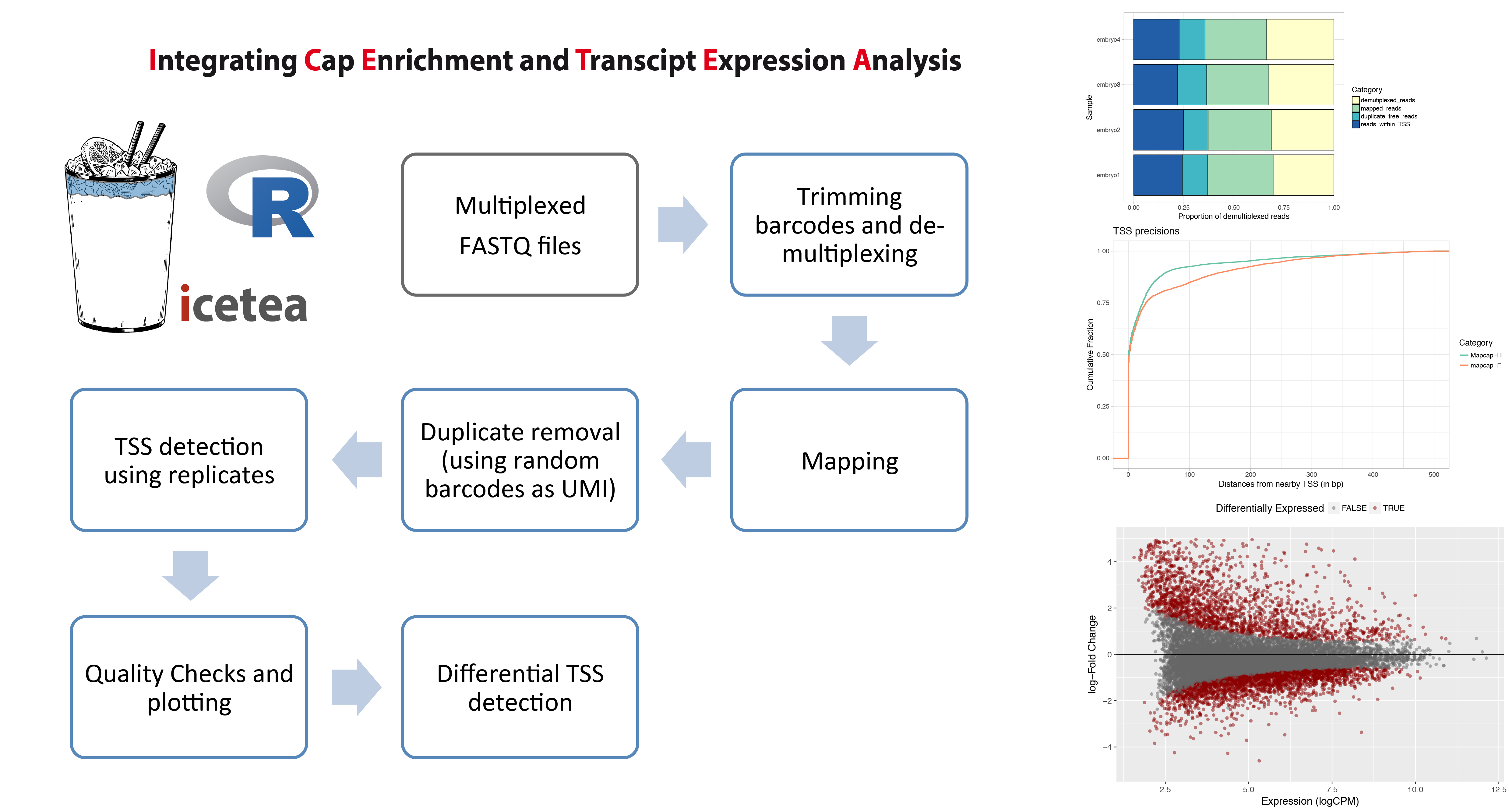
**The icetea R package for analysis of TSS profiling data** allows users to processes data from multiplexed,
5-profiling techniques such as RAMPAGE and the recently developed MAPCap protocol. TSS detection and
differential TSS analysis can be performed using replicates on any 5-profiling dataset.
**Left panel :** Typical analysis steps for MAPCap data that can be performed using icetea.
**Right panel :** Showing some of the quality control and visualization outputs from the package.
Proportion of sequencing reads used for each step (Top), comparison of TSS accuracy (w.r.t. annotated TSS)
between samples (middle), and MA-plots from differential TSS analysis (Bottom).
Additionally, analysis of RNA-binding protein locations via the [FLASH protocol](https://academic.oup.com/nar/article/48/3/e15/5658447) can also be preformed via icetea.
## Installing icetea
### Stable version
Stable release of icetea is available via [bioconductor](https://bioconductor.org/packages/release/bioc/html/icetea.html).
```{r}
## first install BiocManager
install.packages("BiocManager")
## then install icetea
BiocManager::install("icetea")
```
### Latest version
The latest (development) version of icetea can be installed using Devtools. Devtools can be installed from CRAN.
```{r}
## first install devtools
install.packages("devtools")
## then install icetea
devtools::install_github("vivekbhr/icetea")
```
## Documentation
Please visit the icetea [website](https://vivekbhr.github.io/icetea/) for the package documentation and vignette.
## Citation
If you use icetea in your analysis, please cite:
Bhardwaj, V., Semplicio, G., Erdogdu, N. U., Manke, T. & Akhtar, A. MAPCap allows high-resolution detection and differential expression analysis of transcription start sites. [Nat. Commun. 10, 3219 (2019)](https://www.nature.com/articles/s41467-019-11115-x)
Owner
- Name: Vivek Bhardwaj
- Login: vivekbhr
- Kind: user
- Location: the Netherlands
- Company: Hubrecht Institute
- Website: https://vivekbhr.github.io
- Twitter: vivekbhr
- Repositories: 6
- Profile: https://github.com/vivekbhr
Computational Biology Researcher at Hubrecht Institute, the Netherlands.
GitHub Events
Total
Last Year
Committers
Last synced: over 2 years ago
Top Committers
| Name | Commits | |
|---|---|---|
| vivekbhr | b****j@i****e | 407 |
| Nitesh Turaga | n****a@g****m | 6 |
| vivekbhr | v****j@h****u | 5 |
| Vivek Bhardwaj | v****r | 5 |
| vobencha | v****a@g****m | 2 |
| Kayla-Morrell | k****l@r****g | 1 |
Committer Domains (Top 20 + Academic)
Packages
- Total packages: 1
-
Total downloads:
- bioconductor 11,249 total
- Total dependent packages: 0
- Total dependent repositories: 0
- Total versions: 5
- Total maintainers: 1
bioconductor.org: icetea
Integrating Cap Enrichment with Transcript Expression Analysis
- Homepage: https://github.com/vivekbhr/icetea
- Documentation: https://bioconductor.org/packages/release/bioc/vignettes/icetea/inst/doc/icetea.pdf
- License: GPL-3 + file LICENSE
-
Latest release: 1.26.0
published 10 months ago
Rankings
Dependent repos count: 0.0%
Dependent packages count: 0.0%
Average: 23.7%
Downloads: 71.0%
Maintainers (1)
Last synced:
6 months ago
Dependencies
DESCRIPTION
cran
- R >= 4.0 depends
- BiocGenerics * imports
- BiocParallel * imports
- Biostrings * imports
- DESeq2 * imports
- GenomicAlignments * imports
- GenomicFeatures * imports
- GenomicRanges * imports
- IRanges * imports
- Rsamtools * imports
- S4Vectors * imports
- ShortRead * imports
- SummarizedExperiment * imports
- TxDb.Dmelanogaster.UCSC.dm6.ensGene * imports
- VariantAnnotation * imports
- csaw * imports
- edgeR * imports
- ggplot2 * imports
- grDevices * imports
- graphics * imports
- limma * imports
- methods * imports
- rtracklayer * imports
- stats * imports
- utils * imports
- Rsubread >= 1.29.0 suggests
- knitr * suggests
- rmarkdown * suggests
- testthat * suggests